You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_72567-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_72567-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_72567-t26_1-p1 | |||||||||||
| CAZy Family | GT109 | |||||||||||
| CAZyme Description | glycoside hydrolase family 6 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.91:55 | 3.2.1.4:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 139 | 430 | 9.5e-93 | 0.9387755102040817 |
| CBM1 | 31 | 58 | 8.1e-16 | 0.9310344827586207 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 4.80e-164 | 153 | 434 | 13 | 293 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 6.76e-47 | 115 | 469 | 24 | 442 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| 197593 | fCBD | 8.12e-15 | 30 | 62 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 1.31e-14 | 31 | 58 | 2 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 471 | 1 | 471 | |
| 0.0 | 1 | 471 | 1 | 471 | |
| 0.0 | 1 | 471 | 1 | 471 | |
| 0.0 | 1 | 471 | 1 | 471 | |
| 0.0 | 1 | 471 | 1 | 471 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-276 | 107 | 471 | 1 | 365 | Chain A, CELLOBIOHYDROLASE II CORE PROTEIN [Trichoderma reesei] |
|
| 5.28e-276 | 107 | 471 | 1 | 365 | Chain A, CELLOBIOHYDROLASE II [Trichoderma reesei],1CB2_B Chain B, CELLOBIOHYDROLASE II [Trichoderma reesei],1QJW_A Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QJW_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei] |
|
| 2.15e-275 | 107 | 471 | 1 | 365 | Chain A, Cellobiohydrolase Cel6a (formerly Called Cbh Ii) [Trichoderma reesei],1HGY_B Chain B, Cellobiohydrolase Cel6a (formerly Called Cbh Ii) [Trichoderma reesei] |
|
| 2.15e-275 | 107 | 471 | 1 | 365 | Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1HGW_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei] |
|
| 2.83e-275 | 109 | 471 | 1 | 363 | Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK0_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK2_A Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK2_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 471 | 1 | 471 | Exoglucanase 2 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=cbh2 PE=1 SV=1 |
|
| 0.0 | 1 | 471 | 1 | 471 | Exoglucanase 2 OS=Hypocrea jecorina OX=51453 GN=cbh2 PE=1 SV=1 |
|
| 1.29e-205 | 25 | 470 | 17 | 459 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=cbhC PE=3 SV=1 |
|
| 4.95e-202 | 15 | 470 | 8 | 455 | 1,4-beta-D-glucan cellobiohydrolase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cbhC PE=1 SV=2 |
|
| 4.22e-200 | 9 | 471 | 7 | 462 | Putative endoglucanase type B OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
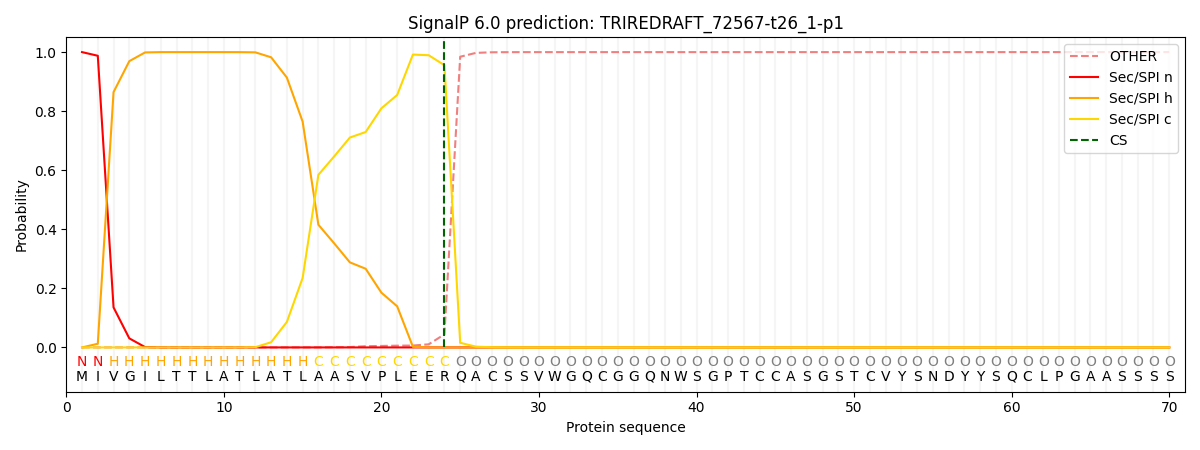
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000251 | 0.999714 | CS pos: 24-25. Pr: 0.9556 |
