You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_68347-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_68347-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_68347-t26_1-p1 | |||||||||||
| CAZy Family | GH75 | |||||||||||
| CAZyme Description | glycoside hydrolase family 18, chitinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:1 | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 34 | 322 | 3.3e-22 | 0.9628378378378378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 2.98e-96 | 32 | 328 | 1 | 280 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 395573 | Glyco_hydro_18 | 9.01e-13 | 35 | 284 | 3 | 239 | Glycosyl hydrolases family 18. |
| 395595 | CBM_1 | 3.60e-10 | 369 | 397 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 6.37e-10 | 368 | 401 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 119350 | GH18_chitinase_D-like | 3.80e-07 | 37 | 318 | 6 | 308 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.14e-246 | 1 | 401 | 1 | 401 | |
| 1.14e-246 | 1 | 401 | 1 | 401 | |
| 3.85e-238 | 1 | 401 | 1 | 401 | |
| 6.15e-220 | 10 | 401 | 10 | 416 | |
| 7.54e-213 | 1 | 401 | 1 | 412 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.11e-57 | 35 | 331 | 4 | 308 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 3.20e-57 | 35 | 331 | 5 | 309 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 3.20e-57 | 35 | 331 | 5 | 309 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 1.72e-55 | 35 | 332 | 9 | 289 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 2.69e-45 | 32 | 308 | 1 | 254 | cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity [Parkia platycephala] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.58e-167 | 7 | 401 | 7 | 419 | Endochitinase 2 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi2 PE=3 SV=2 |
|
| 5.25e-165 | 7 | 401 | 7 | 419 | Endochitinase 2 OS=Metarhizium anisopliae OX=5530 GN=chi2 PE=1 SV=1 |
|
| 4.51e-53 | 35 | 331 | 31 | 337 | Endochitinase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=ctcA PE=2 SV=1 |
|
| 1.30e-52 | 35 | 331 | 32 | 336 | Endochitinase A1 OS=Neosartorya fumigata OX=746128 GN=chiA1 PE=1 SV=1 |
|
| 3.85e-52 | 35 | 332 | 30 | 310 | Endochitinase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CTS1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
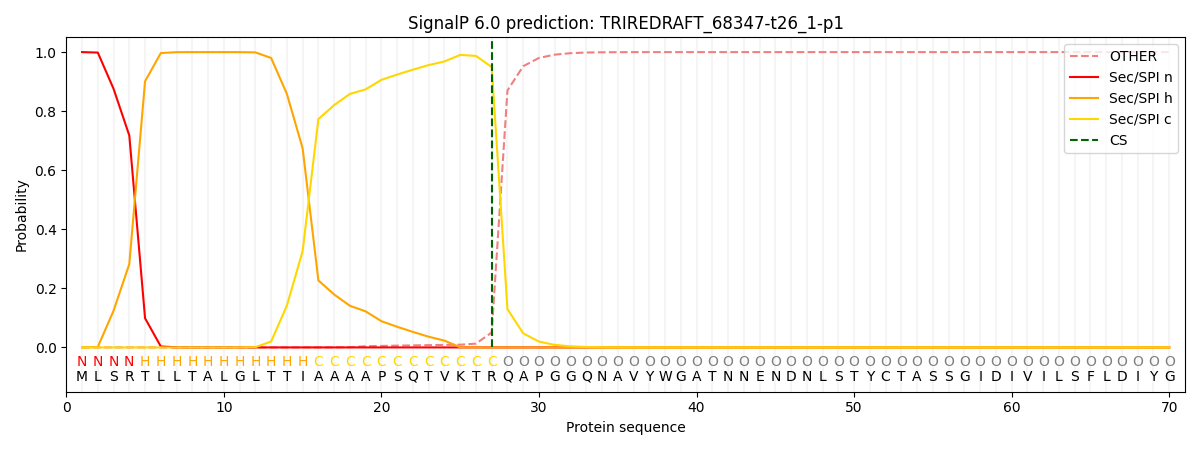
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000293 | 0.999683 | CS pos: 27-28. Pr: 0.9495 |
