You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_49976-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_49976-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_49976-t26_1-p1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | glycoside hydrolase family 45 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 409015 | DPBB_GH45_endoglucanase | 6.94e-33 | 20 | 155 | 7 | 149 | double-psi beta-barrel fold of glycoside hydrolase family 45 endoglucanase EG27II and similar proteins. This group is made up of endoglucanases from mollusks similar to Ampullaria crossean endoglucanase EG27II, a glycoside hydrolase family 45 (GH45) subfamily B protein. Endoglucanases (EC 3.2.1.4) catalyze the endohydrolysis of (1-4)-beta-D-glucosidic linkages in cellulose, lichenin, and cereal beta-D-glucans. Animal cellulases, such as endoglucanase EG27II, have great potential for industrial applications such as bioethanol production. GH45 endoglucanases from mollusks adopt a double-psi beta-barrel (DPBB) fold. |
| 395595 | CBM_1 | 1.34e-09 | 209 | 237 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 3.19e-08 | 208 | 241 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 409011 | DPBB_EXPA_N | 3.62e-08 | 19 | 118 | 4 | 95 | N-terminal double-psi beta-barrel fold domain of the alpha-expansin subfamily. Alpha-expansins (EXPA, expansin-A) have cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. They also affect environmental stress responses. Arabidopsis thaliana EXPA1 is a cell wall modifying enzyme that controls the divisions marking lateral root initiation. Nicotiana tabacum EXPA4 positively regulates abiotic stress tolerance, and negatively regulates pathogen resistance. Wheat TaEXPA2 is involved in conferring cadmium tolerance. Alpha-expansins belong to the expansin family of proteins that contain an N-terminal domain (D1) homologous to the catalytic domain of glycoside hydrolase family 45 (GH45) proteins but with no hydrolytic activity, and a C-terminal domain (D2) homologous to group-2 grass pollen allergens. This model represents the N-terminal domain of alpha-expansins, which adopts a double-psi beta-barrel (DPBB) fold. |
| 409008 | DPBB_EXP_N-like | 1.15e-07 | 22 | 135 | 4 | 92 | N-terminal double-psi beta-barrel fold domain of the expansin family and similar domains. The plant expansin family consists of four subfamilies, alpha-expansin (EXPA), beta-expansin (EXPB), expansin-like A (EXLA), and expansin-like B (EXLB). EXPA and EXPB display cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. EXPA proteins function more efficiently on dicotyledonous cell walls, whereas EXPB proteins exhibit specificity for the cell walls of monocotyledons. Expansins also affect environmental stress responses. Expansin family proteins contain an N-terminal domain (D1) homologous to the catalytic domain of glycoside hydrolase family 45 (GH45) proteins but with no hydrolytic activity, and a C-terminal domain (D2) homologous to group-2 grass pollen allergens. This family also includes GH45 endoglucanases from mollusks. This model represents the N-terminal domain of expansins and similar proteins, which adopts a double-psi beta-barrel (DPBB) fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.10e-171 | 1 | 242 | 1 | 242 | |
| 1.10e-171 | 1 | 242 | 1 | 242 | |
| 1.32e-171 | 1 | 242 | 1 | 242 | |
| 7.39e-142 | 1 | 241 | 1 | 238 | |
| 1.54e-136 | 1 | 241 | 1 | 247 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.92e-30 | 20 | 168 | 25 | 180 | Crystal structure of GH45 endoglucanase EG27II in apo-form [Ampullaria crossean],5XBX_A Crystal structure of GH45 endoglucanase EG27II in complex with cellobiose [Ampullaria crossean],5XC4_A Crystal structure of GH45 endoglucanase EG27II at pH4.0, in complex with cellobiose [Ampullaria crossean],5XC8_A Crystal structure of GH45 endoglucanase EG27II at pH5.5, in complex with cellobiose [Ampullaria crossean],5XC9_A Crystal structure of GH45 endoglucanase EG27II at pH8.0, in complex with cellobiose [Ampullaria crossean] |
|
| 4.48e-29 | 20 | 168 | 25 | 180 | Crystal structure of GH45 endoglucanase EG27II D137A mutant in complex with cellobiose [Ampullaria crossean] |
|
| 3.71e-24 | 20 | 171 | 17 | 181 | Chain A, ENDOGLUCANASE [Mytilus edulis] |
|
| 1.47e-09 | 19 | 135 | 12 | 115 | Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],3X2M_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],3X2N_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],5KJO_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],5KJQ_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium] |
|
| 7.14e-09 | 19 | 135 | 12 | 115 | Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],3X2H_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium],3X2I_A Chain A, Endoglucanase V-like protein [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-172 | 1 | 242 | 1 | 242 | Endoglucanase-5 OS=Hypocrea jecorina OX=51453 GN=egl5 PE=3 SV=1 |
|
| 1.91e-23 | 20 | 171 | 17 | 181 | Endoglucanase OS=Mytilus edulis OX=6550 PE=1 SV=1 |
|
| 1.87e-11 | 207 | 241 | 430 | 464 | Endoglucanase 7a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=eg7A PE=1 SV=1 |
|
| 8.27e-11 | 206 | 242 | 20 | 56 | 1,4-beta-D-glucan cellobiohydrolase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cbhC PE=1 SV=2 |
|
| 9.17e-10 | 206 | 242 | 19 | 55 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
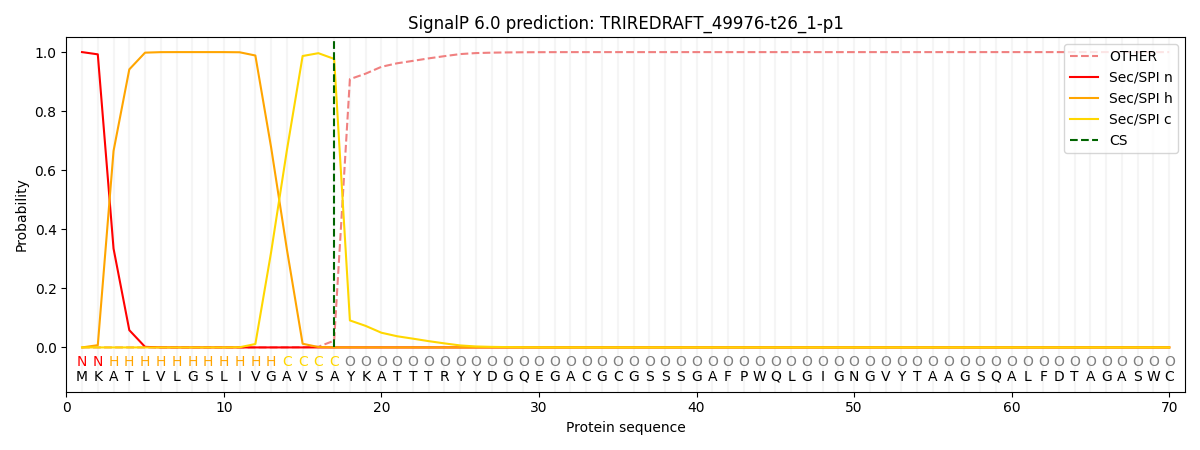
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000271 | 0.999713 | CS pos: 17-18. Pr: 0.9770 |
