You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_122993-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_122993-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_122993-t26_1-p1 | |||||||||||
| CAZy Family | GT64 | |||||||||||
| CAZyme Description | glycosyltransferase family 2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133056 | GT2_HAS | 1.43e-15 | 240 | 421 | 57 | 232 | Hyaluronan synthases catalyze polymerization of hyaluronan. Hyaluronan synthases (HASs) are bi-functional glycosyltransferases that catalyze polymerization of hyaluronan. HASs transfer both GlcUA and GlcNAc in beta-(1,3) and beta-(1,4) linkages, respectively to the hyaluronan chain using UDP-GlcNAc and UDP-GlcUA as substrates. HA is made as a free glycan, not attached to a protein or lipid. HASs do not need a primer for HA synthesis; they initiate HA biosynthesis de novo with only UDP-GlcNAc, UDP-GlcUA, and Mg2+. Hyaluronan (HA) is a linear heteropolysaccharide composed of (1-3)-linked beta-D-GlcUA-beta-D-GlcNAc disaccharide repeats. It can be found in vertebrates and a few microbes and is typically on the cell surface or in the extracellular space, but is also found inside mammalian cells. Hyaluronan has several physiochemical and biological functions such as space filling, lubrication, and providing a hydrated matrix through which cells can migrate. |
| 224136 | BcsA | 1.20e-14 | 275 | 524 | 145 | 368 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 404513 | Glyco_trans_2_3 | 7.59e-08 | 275 | 462 | 6 | 184 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 133043 | CESA_CelA_like | 1.65e-06 | 374 | 419 | 183 | 228 | CESA_CelA_like are involved in the elongation of the glucan chain of cellulose. Family of proteins related to Agrobacterium tumefaciens CelA and Gluconacetobacter xylinus BscA. These proteins are involved in the elongation of the glucan chain of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues. They are putative catalytic subunit of cellulose synthase, which is a glycosyltransferase using UDP-glucose as the substrate. The catalytic subunit is an integral membrane protein with 6 transmembrane segments and it is postulated that the protein is anchored in the membrane at the N-terminal end. |
| 133045 | CESA_like | 2.06e-05 | 275 | 354 | 86 | 165 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 2 | 605 | 1 | 604 | |
| 0.0 | 1 | 605 | 1 | 603 | |
| 0.0 | 1 | 585 | 1 | 583 | |
| 0.0 | 1 | 605 | 1 | 607 | |
| 0.0 | 1 | 605 | 1 | 625 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.76e-22 | 122 | 566 | 89 | 492 | Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2] |
|
| 1.98e-21 | 122 | 566 | 89 | 492 | Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SP8_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SP9_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SPA_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.43e-26 | 122 | 534 | 101 | 493 | Hyaluronan synthase 1 OS=Homo sapiens OX=9606 GN=HAS1 PE=1 SV=2 |
|
| 3.53e-26 | 122 | 534 | 105 | 498 | Hyaluronan synthase 1 OS=Mus musculus OX=10090 GN=Has1 PE=1 SV=1 |
|
| 6.65e-19 | 275 | 458 | 140 | 322 | N-acetylglucosaminyltransferase OS=Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110) OX=224911 GN=nodC PE=3 SV=2 |
|
| 2.49e-18 | 269 | 427 | 133 | 290 | N-acetylglucosaminyltransferase OS=Rhizobium leguminosarum bv. viciae OX=387 GN=nodC PE=3 SV=1 |
|
| 7.29e-18 | 275 | 452 | 140 | 316 | N-acetylglucosaminyltransferase OS=Sinorhizobium fredii (strain NBRC 101917 / NGR234) OX=394 GN=nodC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999975 | 0.000050 |
TMHMM Annotations download full data without filtering help
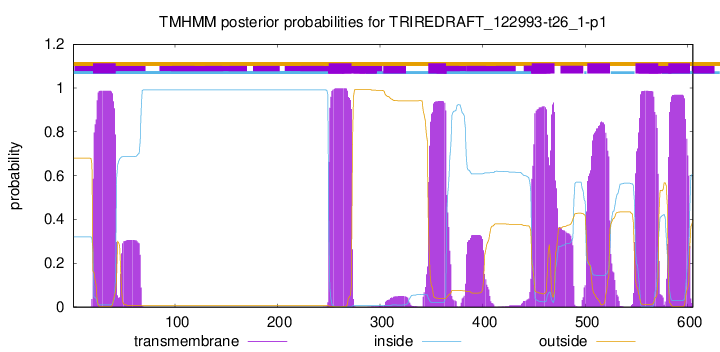
| Start | End |
|---|---|
| 20 | 42 |
| 250 | 272 |
| 347 | 364 |
| 448 | 470 |
| 502 | 524 |
| 549 | 571 |
| 581 | 602 |
