You are browsing environment: FUNGIDB
CAZyme Information: TPX52788.1
You are here: Home > Sequence: TPX52788.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
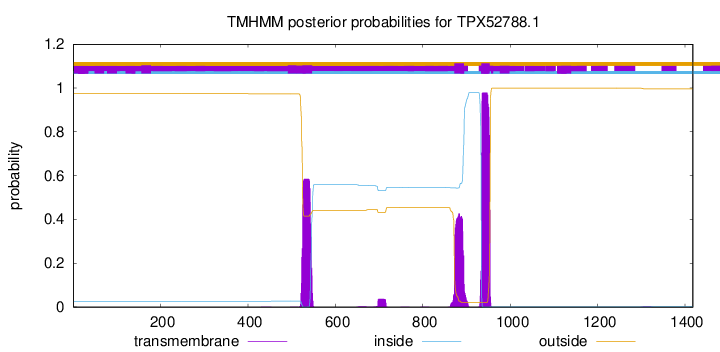
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX52788.1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | DUF1752 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A507DPH5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.131:20 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT15 | 551 | 822 | 3.7e-87 | 0.9926739926739927 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227353 | KTR1 | 5.71e-98 | 550 | 865 | 79 | 388 | Mannosyltransferase [Carbohydrate transport and metabolism]. |
| 396385 | Glyco_transf_15 | 5.68e-88 | 541 | 823 | 30 | 313 | Glycolipid 2-alpha-mannosyltransferase. This is a family of alpha-1,2 mannosyl-transferases involved in N-linked and O-linked glycosylation of proteins. Some of the enzymes in this family have been shown to be involved in O- and N-linked glycan modifications in the Golgi. |
| 400731 | DUF1752 | 7.70e-11 | 33 | 58 | 1 | 26 | Fungal protein of unknown function (DUF1752). This is a family of fungal proteins of unknown function. This short section domain is bounded by two highly conserved tryptophans. The family contains MKD1 that is thought to be a negative regulator of RAS-cAMP pathway in S.cerevisiae. the Sch.pombe member is a GAF1 transcription factor that is also associated with the zinc finger family GATA pfam00320. |
| 396317 | Glyco_transf_92 | 5.78e-06 | 1123 | 1325 | 23 | 210 | Glycosyltransferase family 92. Members of this family act as galactosyltransferases, belonging to glycosyltransferase family 92. The aligned region contains several conserved cysteine residues and several charged residues that may be catalytic residues. This is supported by the inclusion of this family in the GT-A glycosyl transferase superfamily. |
| 404573 | Glyco_tranf_2_4 | 7.46e-06 | 1118 | 1232 | 5 | 90 | Glycosyl transferase family 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.64e-68 | 551 | 845 | 65 | 360 | |
| 2.98e-66 | 551 | 823 | 113 | 382 | |
| 1.17e-64 | 549 | 822 | 47 | 324 | |
| 3.08e-64 | 549 | 842 | 133 | 427 | |
| 3.18e-64 | 546 | 842 | 78 | 377 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.10e-64 | 551 | 842 | 26 | 317 | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p [Saccharomyces cerevisiae],1S4N_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p [Saccharomyces cerevisiae],1S4O_A Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn [Saccharomyces cerevisiae],1S4O_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn [Saccharomyces cerevisiae],1S4P_A Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: ternary complex with GDP/Mn and methyl-alpha-mannoside acceptor [Saccharomyces cerevisiae],1S4P_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: ternary complex with GDP/Mn and methyl-alpha-mannoside acceptor [Saccharomyces cerevisiae] |
|
| 2.05e-58 | 546 | 823 | 98 | 383 | X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae in complex with GDP [Saccharomyces cerevisiae S288C],5A07_B X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae in complex with GDP [Saccharomyces cerevisiae S288C],5A08_A X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae [Saccharomyces cerevisiae S288C],5A08_B X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae [Saccharomyces cerevisiae S288C] |
|
| 3.61e-56 | 551 | 842 | 66 | 361 | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_B Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_C Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_D Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_E Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_F Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOP_A Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_B Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_C Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_D Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_E Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_F Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-62 | 546 | 863 | 134 | 445 | Glycolipid 2-alpha-mannosyltransferase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNT2 PE=3 SV=4 |
|
| 5.51e-62 | 551 | 842 | 120 | 411 | Glycolipid 2-alpha-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=KRE2 PE=1 SV=1 |
|
| 3.16e-61 | 549 | 838 | 83 | 383 | Probable mannosyltransferase KTR3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=KTR3 PE=1 SV=1 |
|
| 4.97e-61 | 547 | 865 | 79 | 413 | O-glycoside alpha-1,2-mannosyltransferase homolog 5 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=omh5 PE=3 SV=2 |
|
| 1.13e-59 | 550 | 823 | 70 | 337 | Probable mannosyltransferase MNT3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNT3 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
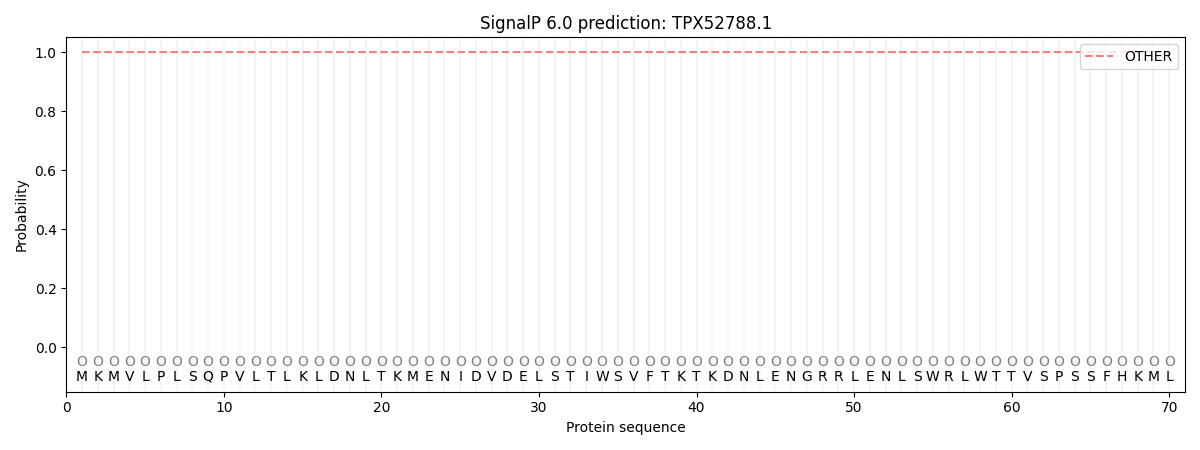
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000023 | 0.000018 |