You are browsing environment: FUNGIDB
CAZyme Information: TPX50495.1
You are here: Home > Sequence: TPX50495.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX50495.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 268 | 395 | 2e-16 | 0.8923076923076924 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200581 | CE4_NodB_like_2 | 6.36e-53 | 274 | 465 | 2 | 189 | Catalytic NodB homology domain of uncharacterized chitin deacetylases and hypothetical proteins. This family includes some uncharacterized chitin deacetylases and hypothetical proteins, mainly from eukaryotes. Although their biological function is unknown, members in this family show high sequence homology to the catalytic NodB homology domain of Colletotrichum lindemuthianum chitin deacetylase (endo-chitin de-N-acetylase, ClCDA, EC 3.5.1.41), which catalyzes the hydrolysis of N-acetamido groups of N-acetyl-D-glucosamine residues in chitin, converting it to chitosan in fungal cell walls. Like ClCDA, this family is a member the carbohydrate esterase 4 (CE4) superfamily. |
| 214618 | RhoGAP | 9.48e-44 | 1140 | 1382 | 1 | 174 | GTPase-activator protein for Rho-like GTPases. GTPase activator proteins towards Rho/Rac/Cdc42-like small GTPases. etter domain limits and outliers. |
| 395495 | RhoGAP | 1.12e-41 | 1143 | 1358 | 1 | 150 | RhoGAP domain. GTPase activator proteins towards Rho/Rac/Cdc42-like small GTPases. |
| 239863 | RhoGAP_fRGD1 | 2.93e-39 | 1142 | 1386 | 16 | 192 | RhoGAP_fRGD1: RhoGAP (GTPase-activator protein [GAP] for Rho-like small GTPases) domain of fungal RGD1-like proteins. Yeast Rgd1 is a GAP protein for Rho3 and Rho4 and plays a role in low-pH response. Small GTPases cluster into distinct families, and all act as molecular switches, active in their GTP-bound form but inactive when GDP-bound. The Rho family of GTPases activates effectors involved in a wide variety of developmental processes, including regulation of cytoskeleton formation, cell proliferation and the JNK signaling pathway. GTPases generally have a low intrinsic GTPase hydrolytic activity but there are family-specific groups of GAPs that enhance the rate of GTP hydrolysis by several orders of magnitude. |
| 238090 | RhoGAP | 1.34e-38 | 1143 | 1380 | 1 | 168 | RhoGAP: GTPase-activator protein (GAP) for Rho-like GTPases; GAPs towards Rho/Rac/Cdc42-like small GTPases. Small GTPases (G proteins) cluster into distinct families, and all act as molecular switches, active in their GTP-bound form but inactive when bound to GDP. The Rho family of small G proteins, which includes Cdc42Hs, activates effectors involved in a wide variety of developmental processes, including regulation of cytoskeleton formation, cell proliferation and the JNK signaling pathway. G proteins generally have a low intrinsic GTPase hydrolytic activity but there are family-specific groups of GAPs that enhance the rate of GTP hydrolysis by several orders of magnitude. The RhoGAPs are one of the major classes of regulators of Rho G proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.10e-34 | 275 | 465 | 73 | 256 | |
| 2.00e-34 | 266 | 446 | 44 | 228 | |
| 6.12e-34 | 275 | 465 | 49 | 233 | |
| 6.12e-34 | 275 | 465 | 49 | 233 | |
| 6.12e-34 | 275 | 465 | 49 | 233 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.31e-22 | 1108 | 1387 | 20 | 230 | Structure of ArhGAP12 bound to G-Actin [Mus musculus],6GVC_R Structure of ArhGAP12 bound to G-Actin [Mus musculus],6GVC_S Structure of ArhGAP12 bound to G-Actin [Mus musculus],6GVC_T Structure of ArhGAP12 bound to G-Actin [Mus musculus] |
|
| 3.10e-21 | 1143 | 1386 | 25 | 202 | The Rho-GAP domain of human N-chimaerin [Homo sapiens] |
|
| 5.66e-21 | 1115 | 1387 | 8 | 212 | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) [Homo sapiens],3BYI_B Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) [Homo sapiens],3BYI_C Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) [Homo sapiens],3BYI_D Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) [Homo sapiens] |
|
| 2.05e-20 | 1143 | 1386 | 289 | 466 | Crystal Structure of the Human Beta2-Chimaerin [Homo sapiens] |
|
| 3.87e-20 | 1145 | 1389 | 40 | 219 | Crystal Structure Of The Bh Domain From Graf, The Gtpase Regulator Associated With Focal Adhesion Kinase [Gallus gallus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.62e-23 | 1109 | 1388 | 657 | 868 | Rho GTPase-activating protein 27 OS=Rattus norvegicus OX=10116 GN=Arhgap27 PE=1 SV=1 |
|
| 4.97e-23 | 1109 | 1388 | 677 | 888 | Rho GTPase-activating protein 27 OS=Homo sapiens OX=9606 GN=ARHGAP27 PE=1 SV=3 |
|
| 6.35e-23 | 1109 | 1388 | 657 | 868 | Rho GTPase-activating protein 27 OS=Mus musculus OX=10090 GN=Arhgap27 PE=1 SV=1 |
|
| 1.10e-21 | 1142 | 1397 | 915 | 1100 | Rho GTPase-activating protein 23 OS=Mus musculus OX=10090 GN=Arhgap23 PE=1 SV=2 |
|
| 4.28e-21 | 1142 | 1397 | 919 | 1104 | Rho GTPase-activating protein 23 OS=Homo sapiens OX=9606 GN=ARHGAP23 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
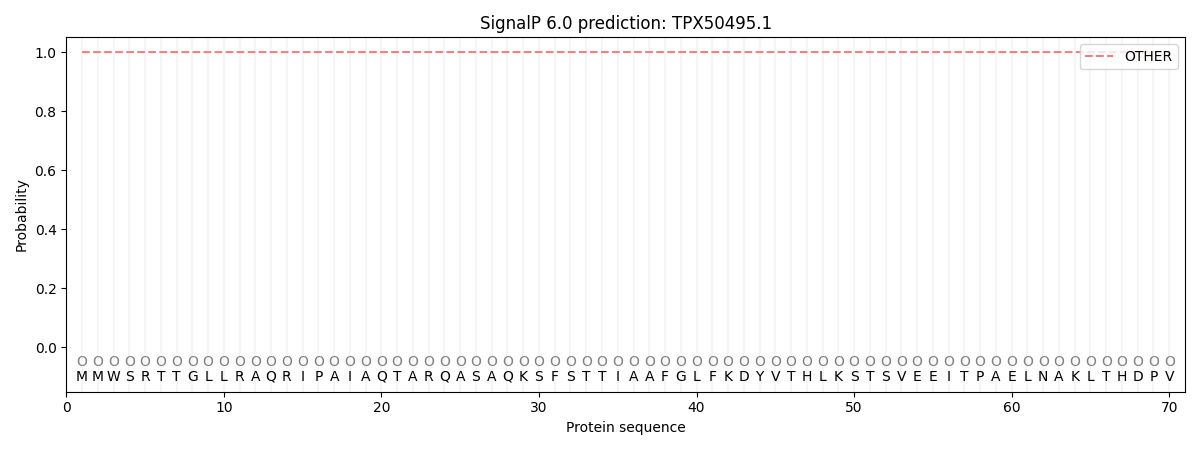
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000000 |
