You are browsing environment: FUNGIDB
CAZyme Information: TPX47790.1
Basic Information
help
| Species |
Synchytrium endobioticum
|
| Lineage |
Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum
|
| CAZyme ID |
TPX47790.1
|
| CAZy Family |
GT14 |
| CAZyme Description |
mannan endo-1,4-beta-mannosidase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 513 |
QEAN01000110|CGC1 |
56294.10 |
5.2684 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_SendobioticumMB42 |
8031 |
N/A |
0 |
8031
|
|
| Gene Location |
No EC number prediction in TPX47790.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
227 |
442 |
1.5e-20 |
0.48514851485148514 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 226609
|
ManB2 |
4.76e-09 |
225 |
469 |
156 |
337 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| 396639
|
Glyco_hydro_26 |
0.003 |
150 |
360 |
55 |
239 |
Glycosyl hydrolase family 26. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 9.22e-60 |
145 |
499 |
47 |
384 |
| 8.00e-56 |
145 |
499 |
47 |
384 |
| 2.13e-55 |
123 |
487 |
26 |
380 |
| 4.24e-50 |
145 |
499 |
52 |
389 |
| 5.91e-50 |
140 |
499 |
48 |
389 |
TPX47790.1 has no PDB hit.
TPX47790.1 has no Swissprot hit.
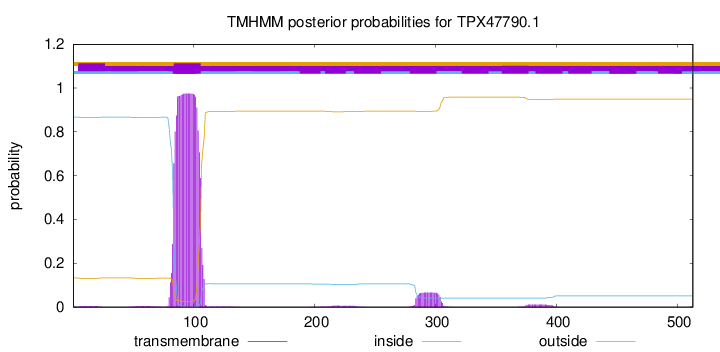
This protein is predicted as OTHER
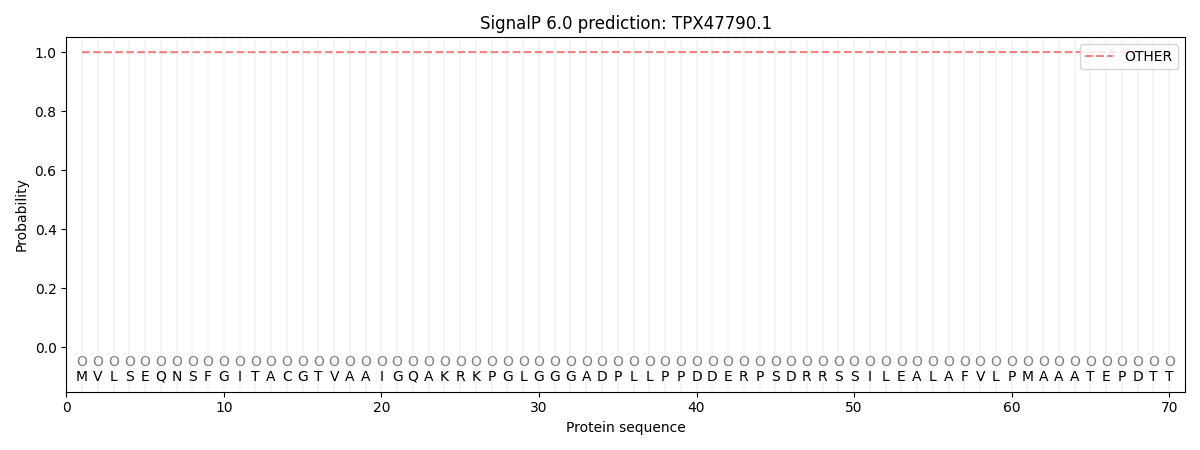
| Other |
SP_Sec_SPI |
CS Position |
| 0.999772 |
0.000268 |
|