You are browsing environment: FUNGIDB
CAZyme Information: TPX46423.1
You are here: Home > Sequence: TPX46423.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX46423.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Ceramide glucosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A507D4M9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33941; End:42766 Strand: + | |||||||||||
Full Sequence Download help
| MPTIDFSIPK RLTQRTSKES IRQLVNGCIE KSETSITLTL APYLASHDRS CSHEHHQYWV | 60 |
| GRDDIAIIPN FLPSPAKAIL DIVLNIRTTS KTRIMFYLTR SNNIIWRGTV LTRAASTIME | 120 |
| VPSCLCIKNV ITSSTHSACS RSGGSEACVA SFGVMMWIIM LQFCLWAAVL IRTRYSNPPL | 180 |
| PTSASTPSAS QSHSTTCKPH PTSPGVTVLR PLKGVDNQLE ENLASAFRQA YPEYQVIMTV | 240 |
| ANRRDPAVAV AKRMIERYPD VDASIVIGNP DLPEVGPNPK VCNLLPGYDL AKHDIIWIVD | 300 |
| SNVILPSIHT LGRSVDALQG PNIGLVHHAP VASHPETFGS RVEMIFLNTA HCKMYLTINY | 360 |
| VAVSSCINGK SNMFRKSDLE RAAKGGLSHF ARYLSEDNIM GEKIWAIGLR HATSGDFAMQ | 420 |
| HLGRSLSLVG YFRRRVRWIR IRKYVVTAAT LIEPFTESIV CGVLALYGFS HWLGWNVKLQ | 480 |
| MLFITSHFMG WLLSDVIIAY TINPSMFSNT SNSGSVVYEC LLYLVAWIVR ECMALPLWVT | 540 |
| GMAGSVIEWR GSLDDRNTFL VYILNRSKMT LNVGKGQHAI MGARKESRKV IPATVDHRFE | 600 |
| SKPALDLVFR QAWEQLNERS EHIFRRHIYI QSIVLTTGNA AGTDTRPDVK TNGADGLPAP | 660 |
| VSAKITPEEL DPNKTDSDPQ GPESKRRVYV NVNPQPPFAH VSHVGNQIRT SKYTLVTFIP | 720 |
| KNVFEQLRSV ANFYFISLVI LQSFDPFKNV DFALTAAPIL FILGVTALKD AFEDYKRHEA | 780 |
| DTRANNTRTY LLSNWKNVNP RPENAAFQFA ALAFYLSSAI SYVFQLLGKP IIRLIRPRML | 840 |
| QESKISNSGD FPPYITFGDY PRCAGDVAQA TYDGVSQDSS SANPNADVAS SADVSGSRVS | 900 |
| NICKSSRGGT SQQLRDSSNA ISSQSSQPKA EKMGTTATTD PVWKLSLWQD AKVGDWVFIR | 960 |
| ENDPIPADVV VVSTSEPDNV CYVETKNLDG ETNLKIRRGI TELAAVKTPA QCAQVKLVID | 1020 |
| SELPSANLYT YSGVATVYPQ ATPASERGAG KVVPIAPVGI LLRGCVLRNT GWIIGIVIYT | 1080 |
| GVETKVMLNS GETPSKRSKI QRTLNPIIVL NFVILVALCL INACAAAIYL GAYRNEGSLW | 1140 |
| NLGDSNSTLT QTILDFFIAM IIYQNIIPIA LYVSVEVAKT IQSVFIYWDD DMYDSEADKH | 1200 |
| VVPHTYNLCD DLGQIEYIFS DKTGTLTANV MEFRMCSING VVYGDGYITE AKIGAMQREG | 1260 |
| VTDFDKGEML SKVAGDEKAM RNALVKDLKA RYIDNKRLSF VDMNLIKDLQ EEEEQARAIR | 1320 |
| EFFILLACCH TLLVDTPKDS AEANTNSNTA KKLIYKAQSP DEAALVAAAR DVGVAFVKRA | 1380 |
| QNSITVDVYS EERTYQVLNV LEFNSDRKRM SVILRDAEGR LVLLCKGADS VIYERLVRGT | 1440 |
| DQALADKTSA DLEGFANDGL RTLCLAWRII PDEVYQSWAE TYLSAQAQIT DREKAIDAAA | 1500 |
| ELIEKDLVLL GATAIEDKLQ DGVPDCIALL AQAGIKIWVL TGDKVETAIN IGFACNLLTG | 1560 |
| TMVLIVVRAT TAKATSDQLK SALNRFWDEN GGAVEGEAHA LIIDGESLKY ALEADCREFL | 1620 |
| LELGCRCKAV VCARVSPLQK ARVVQLVRKG LNVMTLAIGD GANDVSMIQE AEVGVGISGK | 1680 |
| EGLQAVMASD YAIPQFRFLS RLLLVHGKWA YVRTSGLIFN YFYKNIMWLF ILFWYQFESG | 1740 |
| FSADVITDFT YGQFYNTLFT LLPNIYIGTF DQDVNDALSL KLPEIYQKGI RNDLFTNEQV | 1800 |
| WIYIADGIYQ SVIVYWGVSF LFGVDTDPRG YSGNKDAMGT IMAWAAVLIA NFYMAIGTYS | 1860 |
| WTWIAWIALI ATLAMWGAYV TTYAFNVAGL AYGQIPTLYQ QPAFYFVIVL IIIIAFLPRL | 1920 |
| LAATARIMLW PSDTDIVREM QKYDLTGLES TAAKEGRMKL NKDPGPPDTA VDDVGVEVVG | 1980 |
| NEQAVADESG IWAAKRNPVF GAVPPPEPAG APAITIDDVA AIPLTTVNNS NTGATGATLN | 2040 |
| HAGSGQITPP RVVRDSVGNA GQSSNQGDQK PGSDAPKLAV DTTPATLNNG IPTLPAYHPQ | 2100 |
| NDPQAHLRGR TRVSSNRRTS FHLATLPARL VGEFVRSVPR RMRAVSTTHQ DALHTPQRSQ | 2160 |
| GTIVYMENEH VGRIANTGFS FSHEAGMEGV VSPIRVSAGS RSRLRGLQDR APQNSERLSL | 2220 |
| GDSLRKSLHM TPRNRNSSRE PLNVGNTDVT GHPHGNTAGG ETARPSNAGR ERKFSGQKMP | 2280 |
| IRLPFLYNHH NQQQTPYMSE PSLNQSYETH SHRGSIPPGS HSTGIGYGYP PMAPSNDNTS | 2340 |
| NPHQPPPATT ERTVQASSQL AVAPDIEPLT RSPAASNTGT EGQSASGVPA LPLLGQSLTV | 2400 |
| STPSSDGLKS LSPNDGRETT DPRP | 2424 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT21 | 204 | 442 | 2.3e-81 | 0.9957081545064378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273734 | ATPase-Plipid | 0.0 | 704 | 1939 | 2 | 1057 | phospholipid-translocating P-type ATPase, flippase. This model describes the P-type ATPase responsible for transporting phospholipids from one leaflet of bilayer membranes to the other. These ATPases are found only in eukaryotes. |
| 319770 | P-type_ATPase_APLT_Dnf-like | 0.0 | 705 | 1818 | 1 | 836 | Aminophospholipid translocases (APLTs), similar to Saccharomyces cerevisiae Dnf1-3p, Drs2p, and human ATP8A2, -10D, -11B, -11C. Aminophospholipid translocases (APLTs), also known as type 4 P-type ATPases, act as flippases, and translocate specific phospholipids from the exoplasmic leaflet to the cytoplasmic leaflet of biological membranes. Yeast Dnf1 and Dnf2 mediate the transport of phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine from the outer to the inner leaflet of the plasma membrane. This subfamily includes mammalian flippases such as ATP11C which may selectively transports PS and PE from the outer leaflet of the plasma membrane to the inner leaflet. It also includes Arabidopsis phospholipid flippases including ALA1, and Caenorhabditis elegans flippases, including TAT-1, the latter has been shown to facilitate the inward transport of phosphatidylserine. This subfamily belongs to the P-type ATPases, a large family of integral membrane transporters that are of critical importance in all kingdoms of life. They generate and maintain (electro-) chemical gradients across cellular membranes, by translocating cations, heavy metals and lipids, and are distinguished from other main classes of transport ATPases (F- , V- , and ABC- type) by the formation of a phosphorylated (P-) intermediate state in the catalytic cycle. |
| 215623 | PLN03190 | 0.0 | 683 | 1943 | 68 | 1156 | aminophospholipid translocase; Provisional |
| 319838 | P-type_ATPase_APLT | 0.0 | 706 | 1816 | 2 | 805 | Aminophospholipid translocases (APLTs), similar to Saccharomyces cerevisiae Dnf1-3p, Drs2p, Neo1p, and human ATP8A2, -9B, -10D, -11B, and -11C. Aminophospholipid translocases (APLTs), also known as type 4 P-type ATPases, act as flippases, and translocate specific phospholipids from the exoplasmic leaflet to the cytoplasmic leaflet of biological membranes. Yeast Dnf1 and Dnf2 mediate the transport of phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine from the outer to the inner leaflet of the plasma membrane. Mammalian ATP11C may selectively transports PS and PE from the outer leaflet of the plasma membrane to the inner leaflet. The yeast Neo1p localizes to the endoplasmic reticulum and the Golgi complex and plays a role in membrane trafficking within the endomembrane system. Human putative ATPase phospholipid transporting 9B, ATP9B, localizes to the trans-golgi network in a CDC50 protein-independent manner. It also includes Arabidopsis phospholipid flippases including ALA1, and Caenorhabditis elegans flippases, including TAT-1, the latter has been shown to facilitate the inward transport of phosphatidylserine. This subfamily belongs to the P-type ATPases, a large family of integral membrane transporters that are of critical importance in all kingdoms of life. They generate and maintain (electro-) chemical gradients across cellular membranes, by translocating cations, heavy metals and lipids, and are distinguished from other main classes of transport ATPases (F- , V- , and ABC- type) by the formation of a phosphorylated (P-) intermediate state in the catalytic cycle. |
| 223550 | MgtA | 2.05e-135 | 935 | 1960 | 142 | 911 | Magnesium-transporting ATPase (P-type) [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDS10611.1|GT21 | 6.84e-102 | 204 | 551 | 54 | 394 |
| ANB13640.1|GT21 | 9.86e-80 | 203 | 550 | 106 | 475 |
| QRW00262.1|GT21 | 1.07e-79 | 155 | 552 | 16 | 432 |
| QRV86216.1|GT21 | 1.45e-79 | 147 | 552 | 22 | 432 |
| QRW14784.1|GT21 | 4.94e-79 | 147 | 552 | 22 | 432 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LCP_A | 6.23e-269 | 706 | 1939 | 111 | 1340 | Chain A, Phospholipid-transporting ATPase [Thermochaetoides thermophila DSM 1495],6LCR_A Chain A, Phospholipid-transporting ATPase [Thermochaetoides thermophila DSM 1495] |
| 7DRX_A | 7.78e-262 | 706 | 1943 | 194 | 1417 | Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7DSH_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7DSI_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7F7F_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7KY6_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7KYB_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7KYC_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7WHV_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C],7WHW_A Chain A, Phospholipid-transporting ATPase DNF1 [Saccharomyces cerevisiae S288C] |
| 7KY5_A | 8.83e-260 | 706 | 1940 | 232 | 1457 | Chain A, Phospholipid-transporting ATPase DNF2 [Saccharomyces cerevisiae S288C],7KY7_A Chain A, Phospholipid-transporting ATPase DNF2 [Saccharomyces cerevisiae S288C],7KY8_A Chain A, Phospholipid-transporting ATPase DNF2 [Saccharomyces cerevisiae S288C],7KY9_A Chain A, Phospholipid-transporting ATPase DNF2 [Saccharomyces cerevisiae S288C],7KYA_A Chain A, Phospholipid-transporting ATPase DNF2 [Saccharomyces cerevisiae S288C] |
| 6K7G_A | 4.25e-203 | 683 | 1884 | 39 | 1016 | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class1) [Homo sapiens],6K7H_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) [Homo sapiens],6K7I_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) [Homo sapiens],6K7J_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) [Homo sapiens],6K7K_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ADP-Pi state) [Homo sapiens],6K7L_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2P state class2) [Homo sapiens],6K7M_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2Pi-PL state) [Homo sapiens],6K7N_A Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) [Homo sapiens] |
| 6PSY_A | 3.12e-199 | 948 | 1946 | 299 | 1247 | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form [Saccharomyces cerevisiae W303] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q09891|ATCX_SCHPO | 7.06e-275 | 706 | 1939 | 88 | 1295 | Putative phospholipid-transporting ATPase C24B11.12c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC24B11.12c PE=3 SV=1 |
| sp|P32660|ATC5_YEAST | 4.00e-261 | 706 | 1943 | 194 | 1417 | Phospholipid-transporting ATPase DNF1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DNF1 PE=1 SV=2 |
| sp|Q12675|ATC4_YEAST | 4.54e-259 | 706 | 1940 | 232 | 1457 | Phospholipid-transporting ATPase DNF2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DNF2 PE=1 SV=1 |
| sp|O36028|ATCZ_SCHPO | 9.14e-238 | 681 | 1938 | 104 | 1317 | Putative phospholipid-transporting ATPase C4F10.16c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC4F10.16c PE=3 SV=1 |
| sp|Q9XIE6|ALA3_ARATH | 9.48e-217 | 679 | 1966 | 30 | 1151 | Phospholipid-transporting ATPase 3 OS=Arabidopsis thaliana OX=3702 GN=ALA3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000031 | 0.000000 |
TMHMM Annotations download full data without filtering help
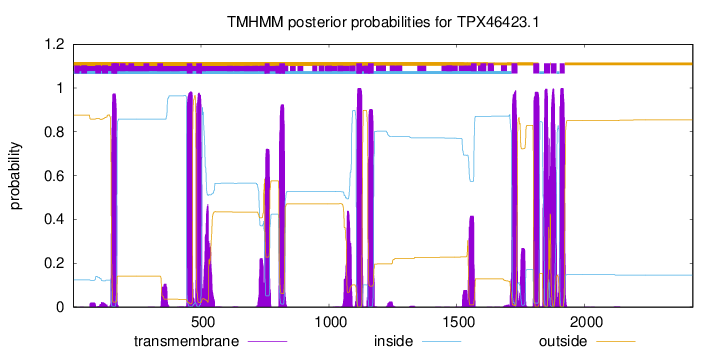
| Start | End |
|---|---|
| 149 | 171 |
| 444 | 466 |
| 481 | 503 |
| 749 | 768 |
| 806 | 828 |
| 1107 | 1129 |
| 1153 | 1175 |
| 1715 | 1737 |
| 1800 | 1822 |
| 1842 | 1864 |
| 1868 | 1890 |
| 1903 | 1922 |
