You are browsing environment: FUNGIDB
CAZyme Information: TPX43706.1
You are here: Home > Sequence: TPX43706.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
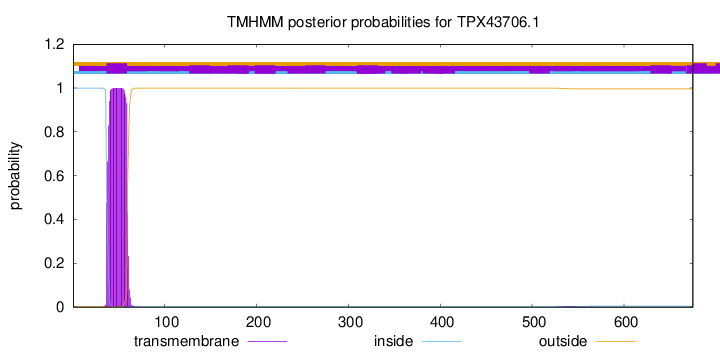
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX43706.1 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.312:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT61 | 473 | 553 | 4.7e-17 | 0.6722689075630253 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398330 | DUF563 | 1.91e-21 | 480 | 581 | 106 | 204 | Protein of unknown function (DUF563). Family of uncharacterized proteins. |
| 226845 | COG4421 | 7.60e-09 | 481 | 562 | 228 | 308 | Capsular polysaccharide biosynthesis protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.15e-58 | 201 | 623 | 68 | 429 | |
| 2.20e-57 | 201 | 623 | 68 | 430 | |
| 2.20e-57 | 201 | 623 | 68 | 430 | |
| 2.93e-57 | 201 | 623 | 68 | 430 | |
| 2.93e-57 | 201 | 623 | 68 | 430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.72e-57 | 201 | 623 | 28 | 391 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9J_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_D Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_E Chain E, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus] |
|
| 5.55e-57 | 201 | 623 | 22 | 385 | Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 9.77e-57 | 201 | 623 | 18 | 381 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 1.06e-56 | 201 | 623 | 22 | 385 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 1.83e-56 | 201 | 623 | 18 | 381 | Chain C, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.91e-58 | 201 | 623 | 68 | 430 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 OS=Xenopus laevis OX=8355 GN=pomgnt2 PE=2 SV=1 |
|
| 5.20e-58 | 201 | 623 | 68 | 430 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 OS=Xenopus tropicalis OX=8364 GN=pomgnt2 PE=2 SV=1 |
|
| 1.88e-56 | 201 | 623 | 68 | 431 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 OS=Danio rerio OX=7955 GN=pomgnt2 PE=2 SV=1 |
|
| 1.95e-56 | 201 | 623 | 69 | 432 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 OS=Bos taurus OX=9913 GN=POMGNT2 PE=1 SV=1 |
|
| 9.76e-56 | 201 | 623 | 69 | 432 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 OS=Rattus norvegicus OX=10116 GN=Pomgnt2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999975 | 0.000035 |