You are browsing environment: FUNGIDB
CAZyme Information: TPX42442.1
You are here: Home > Sequence: TPX42442.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
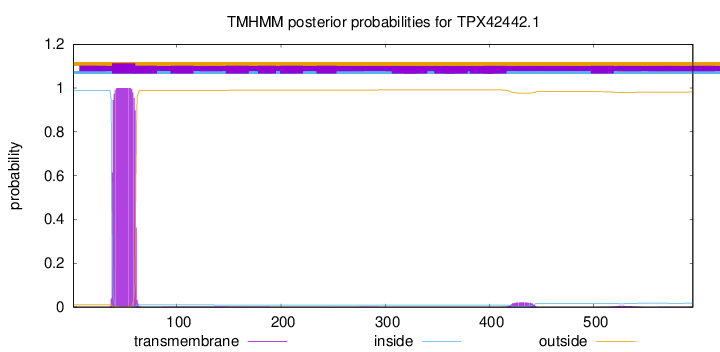
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX42442.1 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Glyco_hydr_30_2 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A507CTE1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 131 | 556 | 4.9e-87 | 0.9298245614035088 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 405300 | Glyco_hydr_30_2 | 3.21e-20 | 139 | 453 | 18 | 367 | O-Glycosyl hydrolase family 30. |
| 227807 | XynC | 1.62e-09 | 239 | 587 | 124 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-61 | 138 | 594 | 48 | 469 | |
| 1.98e-60 | 138 | 594 | 48 | 469 | |
| 5.19e-59 | 138 | 592 | 48 | 467 | |
| 6.39e-59 | 121 | 512 | 21 | 411 | |
| 9.16e-59 | 138 | 593 | 32 | 466 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-07 | 239 | 589 | 88 | 390 | X-ray crystallographic protein structure of the glycoside hydrolase family 30 subfamily 8 xylanase, Xyn30A, from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
|
| 5.19e-06 | 253 | 519 | 97 | 337 | Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.17e-54 | 128 | 512 | 27 | 409 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
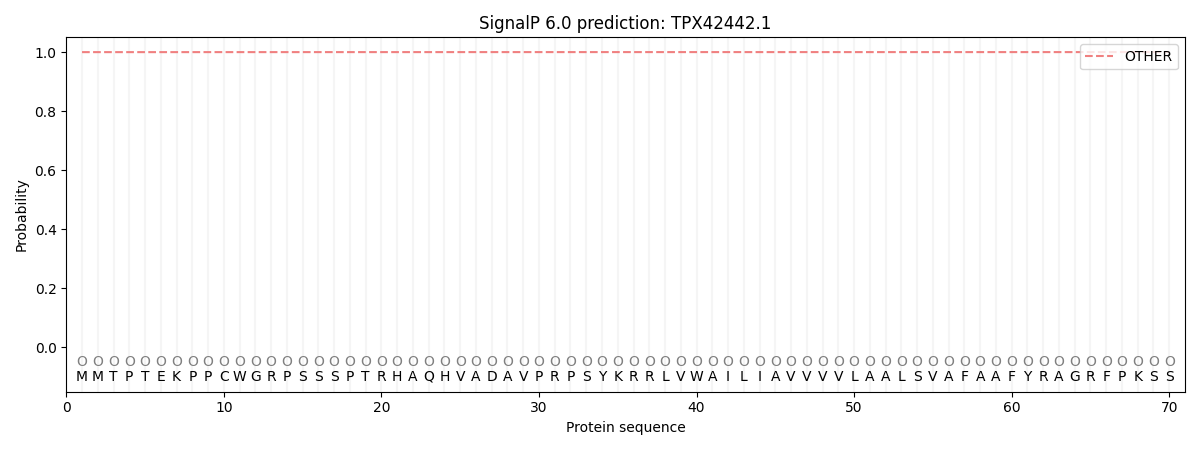
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999971 | 0.000040 |