You are browsing environment: FUNGIDB
CAZyme Information: TPX40007.1
You are here: Home > Sequence: TPX40007.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
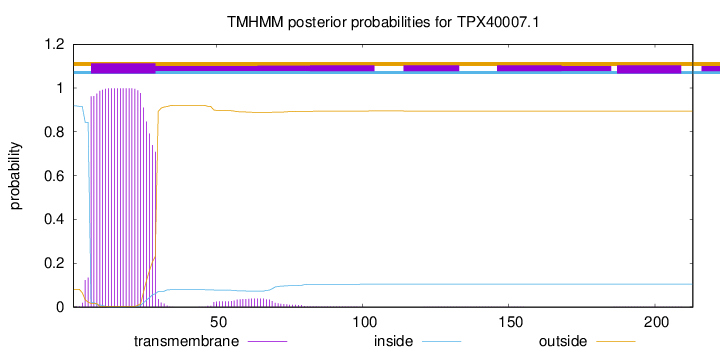
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX40007.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | Cutinase [Source:UniProtKB/TrEMBL;Acc:A0A507CFY6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 43 | 211 | 2e-34 | 0.9947089947089947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 7.04e-40 | 43 | 211 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.99e-45 | 29 | 211 | 49 | 235 | |
| 1.78e-43 | 29 | 211 | 59 | 245 | |
| 6.43e-42 | 35 | 211 | 70 | 250 | |
| 7.09e-42 | 35 | 211 | 36 | 215 | |
| 1.76e-41 | 35 | 211 | 69 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.50e-38 | 39 | 212 | 74 | 248 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 1.15e-21 | 34 | 211 | 6 | 196 | Crystal structure of Aspergillus oryzae cutinase [Aspergillus oryzae] |
|
| 2.46e-21 | 34 | 204 | 12 | 192 | Glomerella cingulata apo cutinase [Colletotrichum gloeosporioides],3DD5_A Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_B Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_C Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_D Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_E Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_F Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_G Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_H Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DEA_A Glomerella cingulata PETFP-cutinase complex [Colletotrichum gloeosporioides],3DEA_B Glomerella cingulata PETFP-cutinase complex [Colletotrichum gloeosporioides] |
|
| 9.77e-21 | 43 | 211 | 11 | 187 | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon [Aspergillus oryzae] |
|
| 6.14e-20 | 33 | 211 | 5 | 192 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.61e-38 | 39 | 212 | 74 | 248 | Cutinase OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=M419DRAFT_76732 PE=1 SV=1 |
|
| 6.61e-38 | 39 | 212 | 74 | 248 | Cutinase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_60489 PE=3 SV=2 |
|
| 8.58e-37 | 44 | 207 | 31 | 196 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 5.23e-35 | 44 | 207 | 31 | 195 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 4.94e-34 | 39 | 212 | 74 | 248 | Cutinase cut1 OS=Trichoderma harzianum OX=5544 GN=cut1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
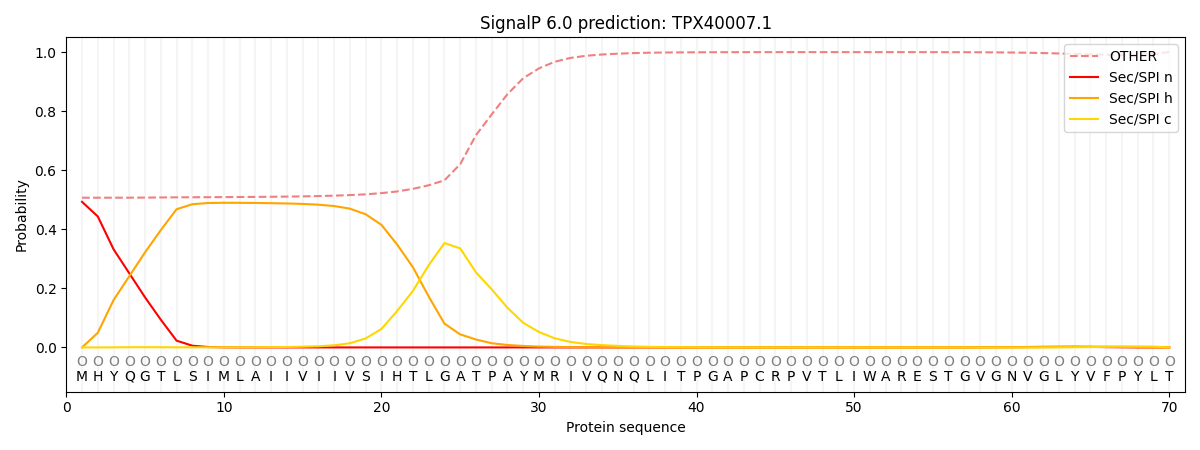
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.520567 | 0.479413 |