You are browsing environment: FUNGIDB
CAZyme Information: TPX39175.1
You are here: Home > Sequence: TPX39175.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX39175.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | 1,3-beta-glucan synthase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 68 | 537 | 2.1e-169 | 0.6211096075778079 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 68 | 594 | 275 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.57e-201 | 41 | 805 | 1123 | 1902 | |
| 2.96e-198 | 68 | 790 | 882 | 1616 | |
| 1.23e-194 | 48 | 763 | 1020 | 1744 | |
| 2.40e-194 | 48 | 763 | 1020 | 1744 | |
| 3.35e-194 | 68 | 763 | 994 | 1702 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.61e-187 | 48 | 787 | 1030 | 1779 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
|
| 1.29e-183 | 68 | 787 | 1114 | 1844 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
|
| 5.22e-173 | 68 | 764 | 998 | 1707 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
|
| 5.18e-172 | 48 | 764 | 1061 | 1795 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
|
| 6.69e-172 | 48 | 764 | 1080 | 1814 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000030 | 0.000002 |
TMHMM Annotations download full data without filtering help
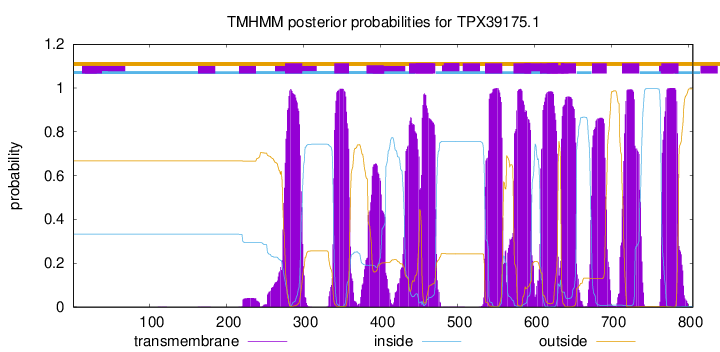
| Start | End |
|---|---|
| 276 | 298 |
| 340 | 359 |
| 437 | 471 |
| 536 | 558 |
| 573 | 595 |
| 608 | 630 |
| 635 | 654 |
| 675 | 694 |
| 714 | 736 |
| 765 | 787 |
