You are browsing environment: FUNGIDB
CAZyme Information: TPX38650.1
You are here: Home > Sequence: TPX38650.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX38650.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | Chitin-binding type-1 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A507CAP3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6863; End:9413 Strand: + | |||||||||||
Full Sequence Download help
| MNLAISILIT TLLASLHTAI AYRCIERYAL TVTTDAQAFG ATAQFDECMH FPNGMALQFQ | 60 |
| SGSLQDLKLP IVISSATCDG NELMQFADLS HNAHKTAYGT YDLLIPLNHS VYVKKIQLRA | 120 |
| QKVDATRLYL DKIFIVQCTP AFHLQKRASE GSCGPEIGSC PFGYCCSTYG YCGTTRAYCG | 180 |
| AGCQANHGNC TVVTIQTSPT VRSSASTANL SPSTSVTFSR SSSITRSISI KPSPFASPTP | 240 |
| SQITSNLPSS SPATEASSII SSRSPPPAVQ NSATRYSTIT APRSTSLTRR PTGANPSPSK | 300 |
| TIKTPTSWDN SEAAFPTLGA SCSSTCGSPQ IQCIDSICQR YWTQPLWTST TLTDKIMLEG | 360 |
| WNVTKYAYDA GRSVTTDPAG GSELVMRVPY PAGSRNPAHE PIGGTGFYAS PLGDMTGMTH | 420 |
| VVMQFDVYFP AEYNFVKGGK LPGGIGGHSG CSGSATATDC YSTRHMWRTN GLGEVYLYIY | 480 |
| QPGQLPGFCS ENNTICIPTD GISVGRGSFK LATGVWNTIK QIVTLNSFDS KGNPVPDDKK | 540 |
| AVIFRLTPDM HPVGIDFETF FGGSDDTWSS PTLQYSYFRR VSLSAY | 586 |
Enzyme Prediction help
| EC | 4.2.2.26:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL14 | 384 | 582 | 4.2e-41 | 0.9809523809523809 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395135 | Chitin_bind_1 | 1.59e-12 | 152 | 185 | 1 | 36 | Chitin recognition protein. |
| 211311 | ChtBD1 | 3.51e-11 | 152 | 186 | 1 | 37 | Hevein or type 1 chitin binding domain. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 211316 | ChtBD1_1 | 3.87e-10 | 152 | 190 | 1 | 43 | Hevein or type 1 chitin binding domain; filamentous ascomycete subfamily. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 214593 | ChtBD1 | 1.79e-09 | 153 | 187 | 2 | 38 | Chitin binding domain. |
| 211312 | ChtBD1_GH19_hevein | 4.14e-08 | 153 | 188 | 3 | 40 | Hevein or Type 1 chitin binding domain subfamily co-occuring with family 19 glycosyl hydrolases or with barwin domains. This subfamily includes Hevein, a major IgE-binding allergen in natural rubber latex. ChtBD1 is a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDS02791.1|PL14_3 | 1.73e-53 | 360 | 586 | 54 | 288 |
| CDS05384.1|PL14_3 | 1.38e-50 | 369 | 583 | 64 | 286 |
| BAE81787.1|PL14|4.2.2.26 | 3.05e-39 | 363 | 584 | 39 | 263 |
| UOH83040.1|PL14_3 | 4.08e-39 | 381 | 583 | 135 | 349 |
| AFQ98373.1|PL14 | 7.85e-39 | 346 | 584 | 19 | 262 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GMT_A | 1.94e-29 | 373 | 583 | 46 | 270 | Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai] |
| 3A0N_A | 1.54e-14 | 350 | 586 | 5 | 243 | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_A Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_B Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3IM0_A Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) [Paramecium bursaria Chlorella virus CVK2] |
SignalP and Lipop Annotations help
This protein is predicted as SP
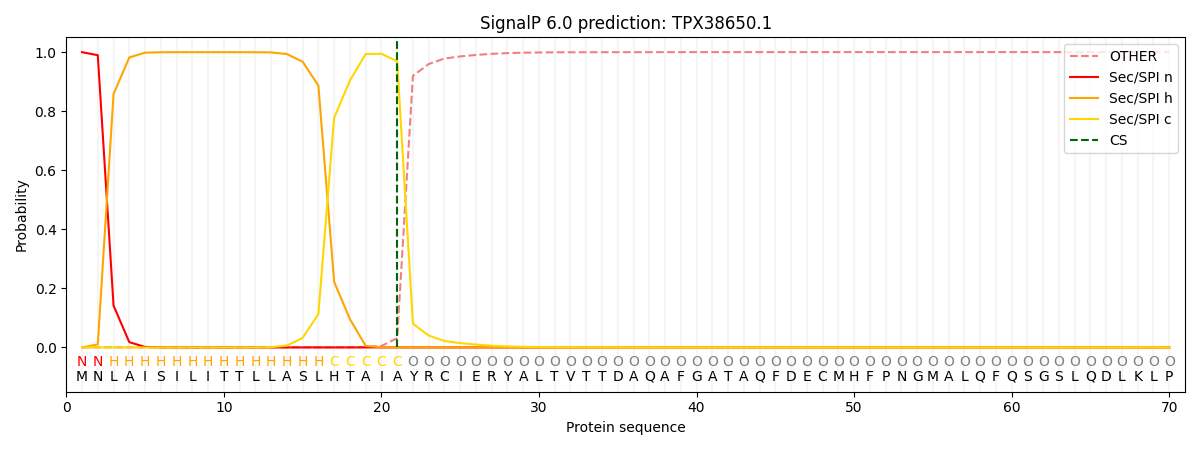
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002136 | 0.997837 | CS pos: 21-22. Pr: 0.9688 |
