You are browsing environment: FUNGIDB
CAZyme Information: TPX36869.1
You are here: Home > Sequence: TPX36869.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX36869.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Fibronectin type-III domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A507CCG5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 116 | 399 | 2.8e-64 | 0.98828125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 2.47e-11 | 162 | 403 | 66 | 295 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 238020 | FN3 | 4.90e-05 | 452 | 535 | 1 | 86 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| 395098 | Cellulase | 8.86e-04 | 215 | 395 | 93 | 272 | Cellulase (glycosyl hydrolase family 5). |
| 394996 | fn3 | 0.003 | 452 | 535 | 1 | 85 | Fibronectin type III domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.69e-82 | 70 | 541 | 28 | 469 | |
| 4.08e-77 | 47 | 541 | 8 | 469 | |
| 4.08e-77 | 47 | 541 | 8 | 469 | |
| 8.27e-75 | 70 | 541 | 28 | 468 | |
| 3.05e-74 | 64 | 541 | 41 | 486 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.63e-22 | 116 | 389 | 47 | 340 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 4.09e-21 | 116 | 389 | 33 | 326 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
|
| 2.98e-18 | 165 | 366 | 89 | 297 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 7.80e-17 | 116 | 297 | 69 | 257 | Exo-mannosidase from Cellvibrio mixtus [Cellvibrio mixtus],1UZ4_A Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis [Cellvibrio mixtus],7ODJ_AAA Chain AAA, Man5A [Cellvibrio mixtus] |
|
| 1.45e-16 | 154 | 366 | 120 | 341 | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei],4LYP_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.59e-20 | 57 | 397 | 7 | 362 | Mannan endo-1,4-beta-mannosidase 1 OS=Solanum lycopersicum OX=4081 GN=MAN1 PE=1 SV=2 |
|
| 5.58e-18 | 61 | 297 | 20 | 240 | Putative mannan endo-1,4-beta-mannosidase 4 OS=Arabidopsis thaliana OX=3702 GN=MAN4 PE=3 SV=1 |
|
| 1.00e-17 | 61 | 297 | 20 | 240 | Putative mannan endo-1,4-beta-mannosidase P OS=Arabidopsis thaliana OX=3702 GN=MANP PE=5 SV=3 |
|
| 1.19e-17 | 117 | 366 | 78 | 333 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
|
| 3.35e-16 | 70 | 297 | 29 | 236 | Mannan endo-1,4-beta-mannosidase 1 OS=Arabidopsis thaliana OX=3702 GN=MAN1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
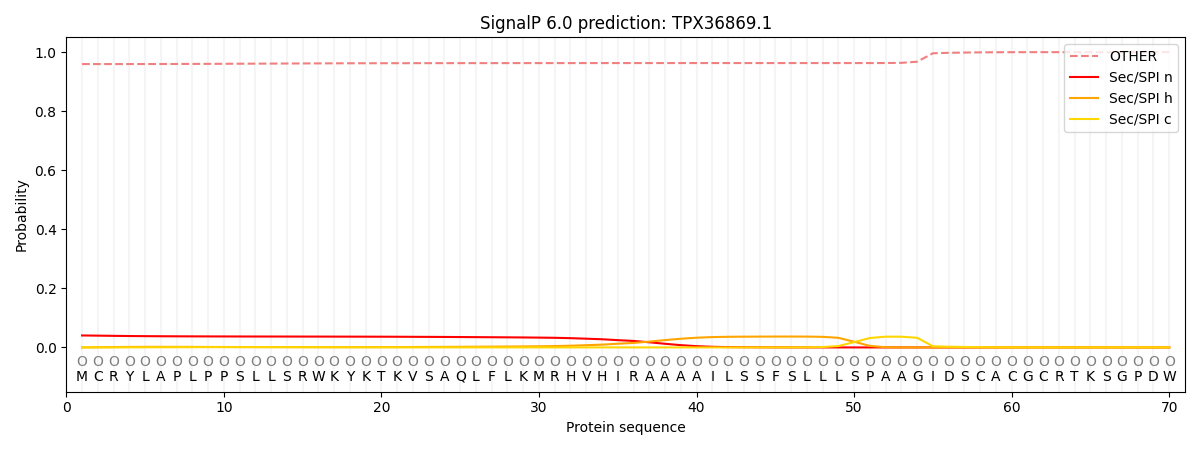
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.961783 | 0.038231 |
