You are browsing environment: FUNGIDB
CAZyme Information: TPX34119.1
You are here: Home > Sequence: TPX34119.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
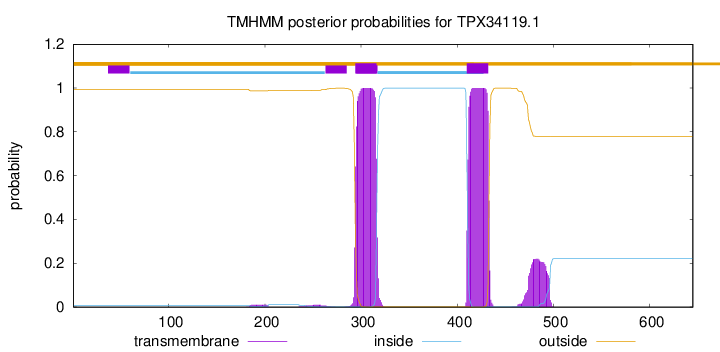
TMHMM annotations
Basic Information help
| Species | Synchytrium endobioticum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; Synchytriaceae; Synchytrium; Synchytrium endobioticum | |||||||||||
| CAZyme ID | TPX34119.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 2.44e-05 | 127 | 295 | 51 | 201 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.30e-61 | 44 | 606 | 27 | 430 | |
| 1.97e-58 | 41 | 606 | 20 | 426 | |
| 1.79e-56 | 38 | 606 | 17 | 427 | |
| 2.14e-55 | 48 | 606 | 27 | 426 | |
| 2.32e-55 | 48 | 606 | 29 | 427 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.12e-13 | 128 | 286 | 71 | 259 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 2.94e-12 | 134 | 291 | 78 | 259 | Chain A, endo-beta-mannanase [Solanum lycopersicum] |
|
| 2.19e-09 | 143 | 270 | 102 | 230 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 2.41e-09 | 140 | 264 | 89 | 207 | Chain A, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
|
| 3.64e-09 | 143 | 270 | 88 | 216 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.22e-14 | 135 | 286 | 111 | 290 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
|
| 3.66e-13 | 134 | 279 | 150 | 306 | Putative mannan endo-1,4-beta-mannosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN5 PE=2 SV=2 |
|
| 2.26e-12 | 134 | 286 | 119 | 300 | Mannan endo-1,4-beta-mannosidase 6 OS=Arabidopsis thaliana OX=3702 GN=MAN6 PE=2 SV=1 |
|
| 1.19e-11 | 140 | 264 | 184 | 302 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
|
| 1.19e-11 | 140 | 264 | 184 | 302 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
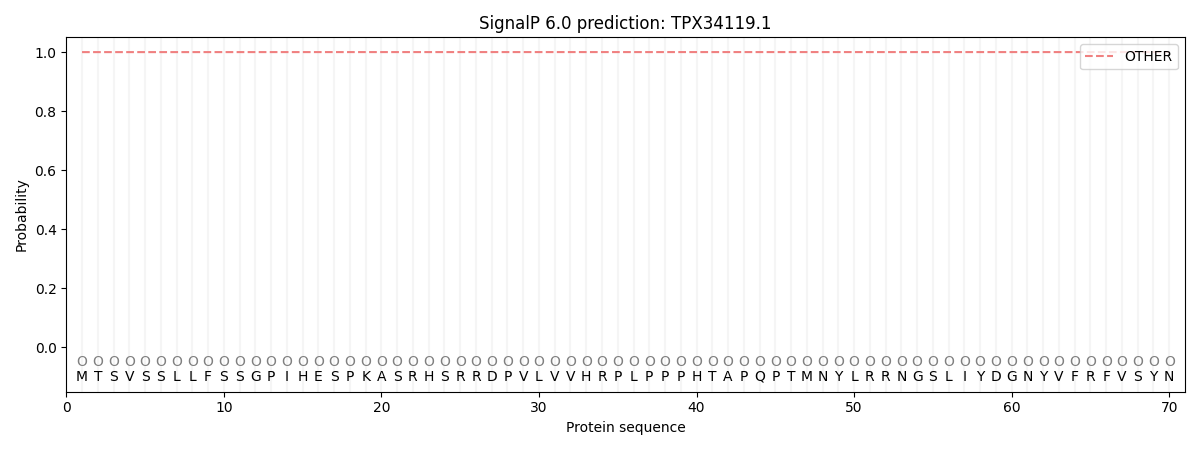
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000021 |