You are browsing environment: FUNGIDB
CAZyme Information: THC98270.1
You are here: Home > Sequence: THC98270.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC98270.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 32 | 367 | 4.1e-92 | 0.9050279329608939 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.84e-32 | 134 | 357 | 105 | 339 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 1.38e-11 | 49 | 370 | 46 | 351 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| 224250 | YyaL | 3.93e-04 | 246 | 322 | 473 | 558 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| 399891 | GlcNAc_2-epim | 0.007 | 170 | 286 | 81 | 186 | N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase). This family contains a number of eukaryotic and bacterial N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase) enzymes (EC:5.3.1.8) approximately 500 residues long. This converts N-acyl-D-glucosamine to N-acyl-D-mannosamine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.82e-170 | 1 | 369 | 1 | 372 | |
| 1.82e-170 | 1 | 369 | 1 | 372 | |
| 1.82e-170 | 1 | 369 | 1 | 372 | |
| 1.82e-170 | 1 | 369 | 1 | 372 | |
| 1.82e-170 | 1 | 369 | 1 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.38e-16 | 107 | 365 | 75 | 338 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 3.47e-16 | 107 | 365 | 78 | 341 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 7.32e-16 | 107 | 365 | 96 | 359 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 1.52e-15 | 107 | 365 | 109 | 372 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
|
| 3.26e-15 | 107 | 365 | 96 | 359 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5AGD_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBM_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.22e-22 | 96 | 316 | 84 | 318 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
|
| 1.04e-15 | 121 | 346 | 122 | 367 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
|
| 1.43e-14 | 121 | 325 | 114 | 334 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
|
| 5.00e-12 | 121 | 328 | 110 | 334 | Putative mannan endo-1,6-alpha-mannosidase C1198.06c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.06c PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
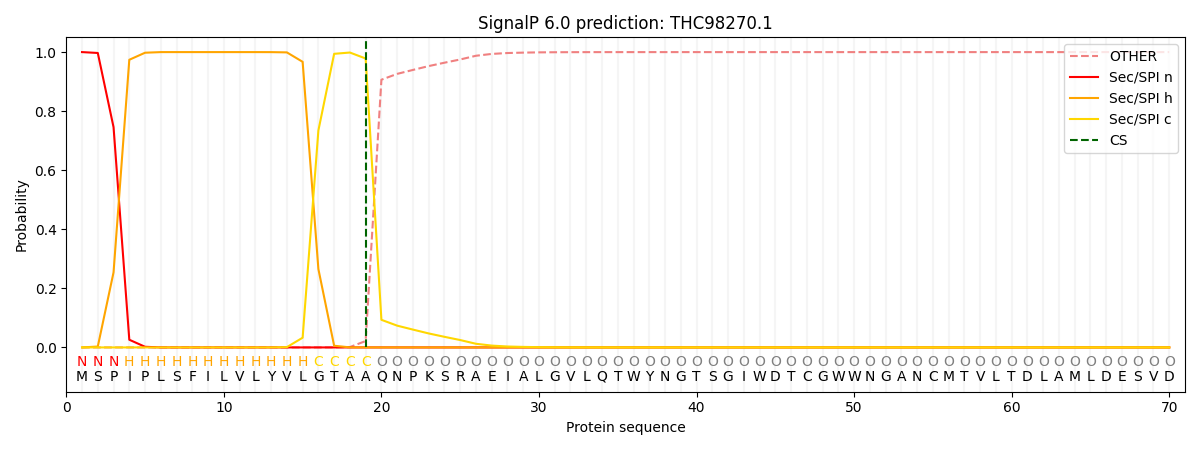
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999743 | CS pos: 19-20. Pr: 0.9783 |
