You are browsing environment: FUNGIDB
CAZyme Information: THC97742.1
You are here: Home > Sequence: THC97742.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC97742.1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | SGNH_hydro domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A4S3JQ19] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 180 | 370 | 6.3e-17 | 0.84688995215311 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238869 | Endoglucanase_E_like | 1.81e-46 | 163 | 373 | 1 | 169 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| 238141 | SGNH_hydrolase | 1.28e-16 | 164 | 370 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 404371 | Lipase_GDSL_2 | 8.94e-10 | 189 | 361 | 8 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.36e-202 | 15 | 399 | 14 | 402 | |
| 1.16e-198 | 14 | 399 | 14 | 407 | |
| 5.28e-193 | 5 | 399 | 6 | 409 | |
| 5.47e-191 | 15 | 399 | 14 | 392 | |
| 2.44e-167 | 1 | 399 | 1 | 398 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
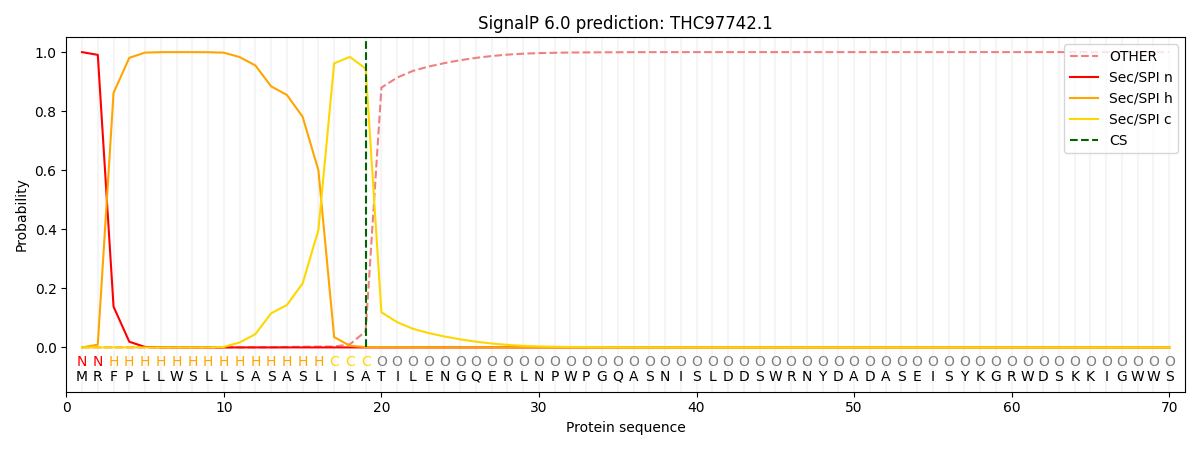
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000684 | 0.999308 | CS pos: 19-20. Pr: 0.9440 |
