You are browsing environment: FUNGIDB
CAZyme Information: THC96448.1
You are here: Home > Sequence: THC96448.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC96448.1 | |||||||||||
| CAZy Family | GH65 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 161 | 610 | 3.2e-101 | 0.9766899766899767 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 1.33e-80 | 162 | 605 | 5 | 442 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 2.97e-78 | 160 | 604 | 2 | 443 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 7.68e-46 | 161 | 592 | 27 | 459 | beta-glucosidase |
| 184975 | PRK15014 | 5.64e-43 | 162 | 604 | 6 | 463 | 6-phospho-beta-glucosidase BglA; Provisional |
| 215455 | PLN02849 | 1.05e-41 | 161 | 592 | 29 | 459 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 615 | 1 | 616 | |
| 0.0 | 57 | 615 | 24 | 581 | |
| 0.0 | 1 | 614 | 1 | 615 | |
| 0.0 | 8 | 614 | 6 | 619 | |
| 1.84e-317 | 1 | 614 | 1 | 615 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.41e-74 | 85 | 603 | 8 | 560 | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis [Hamamotoa singularis] |
|
| 3.26e-73 | 162 | 603 | 89 | 560 | Chain A, Beta-hexosyltransferase [Hamamotoa singularis],7L74_B Chain B, Beta-hexosyltransferase [Hamamotoa singularis] |
|
| 3.26e-73 | 85 | 603 | 8 | 560 | Crystal structure of the E496A mutant of HsBglA [Hamamotoa singularis],6M4F_C Crystal structure of the E496A mutant of HsBglA [Hamamotoa singularis],6M4F_E Crystal structure of the E496A mutant of HsBglA [Hamamotoa singularis],6M4F_G Crystal structure of the E496A mutant of HsBglA [Hamamotoa singularis] |
|
| 1.05e-72 | 162 | 603 | 57 | 528 | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose [Hamamotoa singularis],6M55_D Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose [Hamamotoa singularis] |
|
| 1.41e-67 | 163 | 603 | 10 | 451 | Chain A, Beta-glucosidase [Phanerodontia chrysosporium],2E3Z_B Chain B, Beta-glucosidase [Phanerodontia chrysosporium],2E40_A Chain A, Beta-glucosidase [Phanerodontia chrysosporium],2E40_B Chain B, Beta-glucosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.74e-67 | 163 | 603 | 7 | 448 | Beta-glucosidase 1A OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1A PE=1 SV=1 |
|
| 8.69e-60 | 163 | 603 | 12 | 463 | Beta-glucosidase 1B OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1B PE=1 SV=1 |
|
| 3.49e-57 | 161 | 592 | 14 | 451 | Beta-glucosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU4 PE=2 SV=1 |
|
| 2.08e-55 | 161 | 592 | 20 | 459 | Beta-glucosidase 42 OS=Arabidopsis thaliana OX=3702 GN=BGLU42 PE=2 SV=1 |
|
| 6.09e-55 | 161 | 603 | 37 | 498 | Beta-glucosidase 34 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU34 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
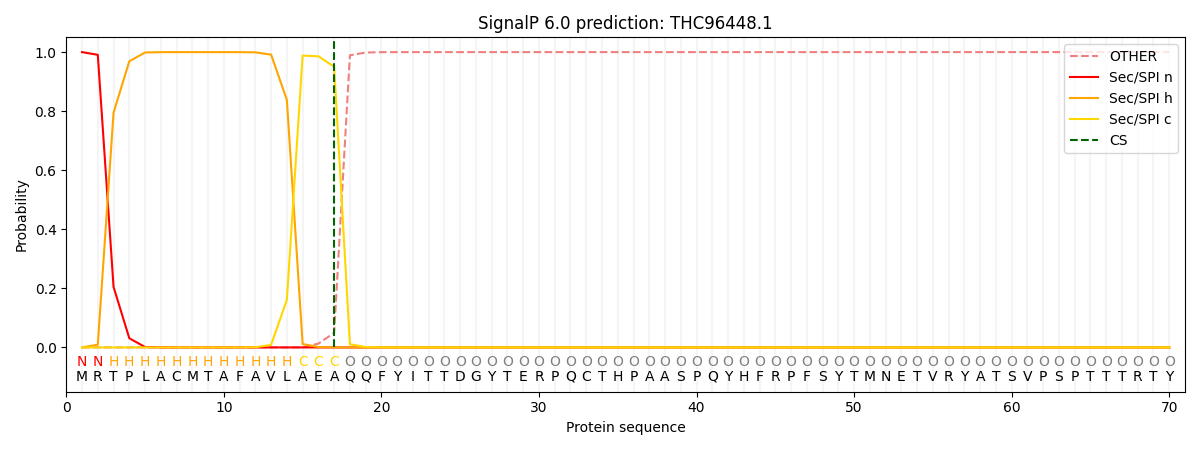
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000245 | 0.999720 | CS pos: 17-18. Pr: 0.9506 |
