You are browsing environment: FUNGIDB
CAZyme Information: THC95862.1
Basic Information
help
| Species |
Aspergillus tanneri
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri
|
| CAZyme ID |
THC95862.1
|
| CAZy Family |
GT90 |
| CAZyme Description |
LysM domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A4S3JKA9]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 375 |
SOSA01000138|CGC1 |
40684.43 |
5.9663 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AtanneriNIH1004 |
13590 |
N/A |
33 |
13557
|
|
| Gene Location |
THC95862.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.06e-187 |
12 |
375 |
11 |
391 |
| 3.06e-187 |
12 |
375 |
11 |
391 |
| 9.54e-147 |
23 |
375 |
21 |
387 |
| 8.83e-145 |
12 |
375 |
10 |
384 |
| 3.63e-140 |
28 |
375 |
29 |
393 |
THC95862.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6.33e-13 |
277 |
373 |
191 |
284 |
Killer toxin subunits alpha/beta OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 PE=1 SV=1 |
This protein is predicted as SP
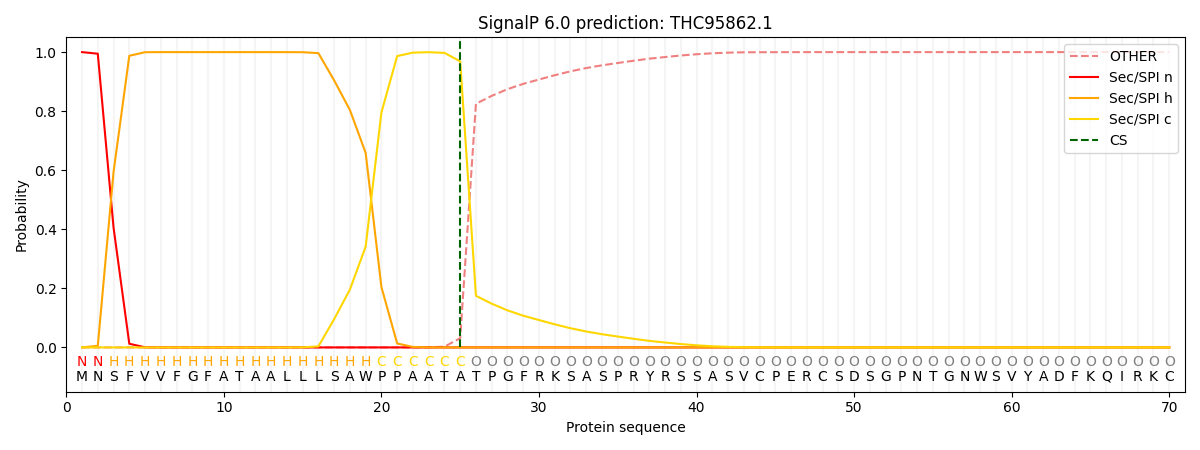
| Other |
SP_Sec_SPI |
CS Position |
| 0.000397 |
0.999558 |
CS pos: 25-26. Pr: 0.9687 |
There is no transmembrane helices in THC95862.1.

