You are browsing environment: FUNGIDB
CAZyme Information: THC92789.1
You are here: Home > Sequence: THC92789.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC92789.1 | |||||||||||
| CAZy Family | GH16|GH16 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 16 | 556 | 7.7e-134 | 0.9911971830985915 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 2.61e-85 | 17 | 558 | 9 | 536 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 1.32e-82 | 17 | 556 | 7 | 532 | choline dehydrogenase; Validated |
| 273814 | betA | 9.28e-70 | 17 | 556 | 1 | 527 | choline dehydrogenase. Choline dehydrogenase catalyzes the conversion of exogenously supplied choline into the intermediate glycine betaine aldehyde, as part of a two-step oxidative reaction leading to the formation of osmoprotectant betaine. This enzymatic system can be found in both gram-positive and gram-negative bacteria. As in Escherichia coli, Staphylococcus xylosus, and Sinorhizobium meliloti, this enzyme is found associated in a transciptionally co-induced gene cluster with betaine aldehyde dehydrogenase, the second catalytic enzyme in this reaction. Other gram-positive organisms have been shown to employ a different enzymatic system, utlizing a soluable choline oxidase or type III alcohol dehydrogenase instead of choline dehydrogenase. This enzyme is a member of the GMC oxidoreductase family (pfam00732 and pfam05199), sharing a common evoluntionary origin and enzymatic reaction with alcohol dehydrogenase. Outgrouping from this model, Caulobacter crescentus shares sequence homology with choline dehydrogenase, yet other genes participating in this enzymatic reaction have not currently been identified. [Cellular processes, Adaptations to atypical conditions] |
| 398739 | GMC_oxred_C | 1.98e-34 | 417 | 551 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 2.36e-21 | 84 | 306 | 14 | 217 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.46e-143 | 7 | 556 | 5 | 608 | |
| 3.79e-142 | 4 | 558 | 2 | 614 | |
| 1.61e-141 | 7 | 558 | 5 | 611 | |
| 6.40e-141 | 11 | 558 | 9 | 611 | |
| 6.40e-141 | 11 | 558 | 9 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.93e-68 | 13 | 561 | 3 | 570 | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase [Aspergillus flavus NRRL3357],4YNU_A Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone [Aspergillus flavus NRRL3357] |
|
| 1.66e-53 | 17 | 558 | 3 | 564 | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid [Pleurotus eryngii] |
|
| 5.91e-53 | 17 | 555 | 42 | 597 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 1.64e-52 | 14 | 558 | 1 | 565 | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii [Pleurotus eryngii] |
|
| 1.26e-50 | 14 | 556 | 4 | 583 | Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE3_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.14e-141 | 11 | 558 | 9 | 611 | Dehydrogenase citC OS=Monascus ruber OX=89489 GN=citC PE=1 SV=1 |
|
| 1.14e-141 | 11 | 558 | 9 | 611 | Dehydrogenase mpl7 OS=Monascus purpureus OX=5098 GN=mpl7 PE=1 SV=1 |
|
| 6.97e-140 | 6 | 558 | 4 | 619 | Dehydrogenase OS=Didymella fabae OX=372025 GN=orf9 PE=3 SV=1 |
|
| 8.11e-73 | 15 | 555 | 46 | 615 | GMC oxidoreductase family protein Mala s 12.0101 OS=Malassezia sympodialis OX=76777 PE=1 SV=2 |
|
| 8.11e-73 | 15 | 555 | 46 | 615 | GMC oxidoreductase family protein Mala s 12 OS=Malassezia sympodialis (strain ATCC 42132) OX=1230383 GN=MSY001_2108 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
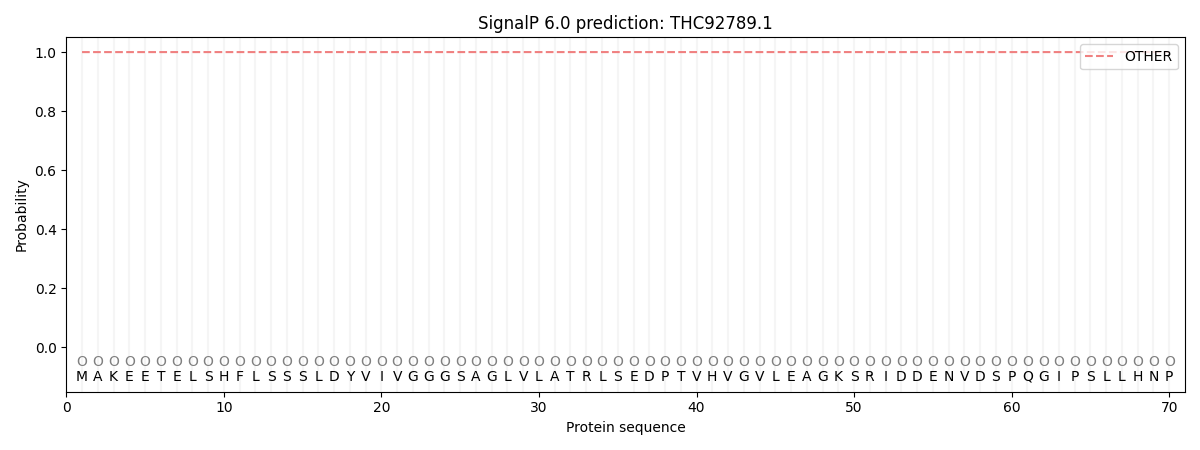
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000048 | 0.000004 |
