You are browsing environment: FUNGIDB
CAZyme Information: THC90992.1
You are here: Home > Sequence: THC90992.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC90992.1 | |||||||||||
| CAZy Family | CE9 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 40 | 597 | 7.8e-142 | 0.9947183098591549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235000 | PRK02106 | 3.42e-56 | 39 | 596 | 4 | 532 | choline dehydrogenase; Validated |
| 225186 | BetA | 9.62e-56 | 35 | 598 | 2 | 536 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 366272 | GMC_oxred_N | 1.78e-41 | 109 | 351 | 16 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 398739 | GMC_oxred_C | 9.25e-29 | 450 | 591 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 215420 | PLN02785 | 2.04e-14 | 41 | 591 | 56 | 576 | Protein HOTHEAD |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 3 | 601 | 5 | 602 | |
| 0.0 | 1 | 599 | 1 | 605 | |
| 0.0 | 7 | 601 | 9 | 603 | |
| 0.0 | 7 | 601 | 9 | 603 | |
| 0.0 | 11 | 599 | 13 | 603 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 17 | 601 | 1 | 585 | Glucose Oxidase From Penicillium Amagasakiense [Penicillium amagasakiense],1GPE_B Glucose Oxidase From Penicillium Amagasakiense [Penicillium amagasakiense] |
|
| 2.82e-293 | 25 | 602 | 2 | 580 | Glucose oxydase mutant A2 [Aspergillus niger] |
|
| 3.03e-293 | 25 | 602 | 4 | 582 | GLUCOSE OXIDASE FROM APERGILLUS NIGER [Aspergillus niger],1GAL_A CRYSTAL STRUCTURE OF GLUCOSE OXIDASE FROM ASPERGILLUS NIGER: REFINED AT 2.3 ANGSTROMS RESOLUTION [Aspergillus niger],3QVP_A Crystal structure of glucose oxidase for space group C2221 at 1.2 A resolution [Aspergillus niger],3QVR_A Crystal structure of glucose oxidase for space group P3121 at 1.3 A resolution. [Aspergillus niger] |
|
| 4.00e-293 | 25 | 602 | 2 | 580 | Glucose oxidase mutant A2 [Aspergillus niger] |
|
| 4.92e-102 | 41 | 598 | 6 | 567 | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase [Aspergillus flavus NRRL3357],4YNU_A Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone [Aspergillus flavus NRRL3357] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 7 | 601 | 9 | 603 | Glucose oxidase OS=Talaromyces flavus OX=5095 GN=GOX PE=3 SV=1 |
|
| 0.0 | 17 | 601 | 1 | 585 | Glucose oxidase OS=Penicillium amagasakiense OX=63559 PE=1 SV=1 |
|
| 2.46e-292 | 3 | 602 | 5 | 604 | Glucose oxidase OS=Aspergillus niger OX=5061 GN=gox PE=1 SV=1 |
|
| 1.57e-71 | 35 | 598 | 41 | 618 | GMC oxidoreductase family protein Mala s 12 OS=Malassezia sympodialis (strain ATCC 42132) OX=1230383 GN=MSY001_2108 PE=1 SV=1 |
|
| 1.57e-71 | 35 | 598 | 41 | 618 | GMC oxidoreductase family protein Mala s 12.0101 OS=Malassezia sympodialis OX=76777 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
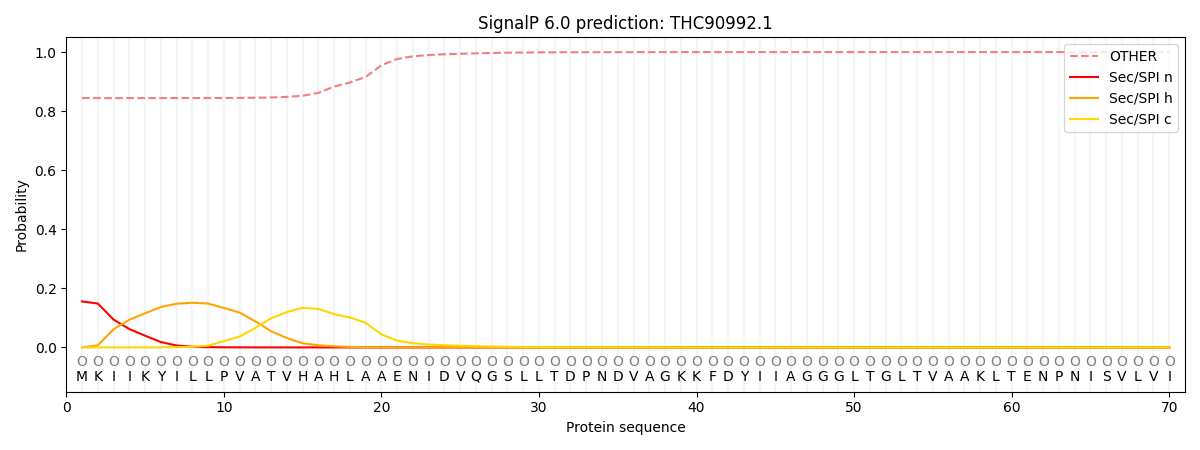
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.849630 | 0.150387 |
