You are browsing environment: FUNGIDB
CAZyme Information: THC88014.1
You are here: Home > Sequence: THC88014.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus tanneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus tanneri | |||||||||||
| CAZyme ID | THC88014.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | NodB homology domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A4V3UMN8] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 46 | 149 | 2.9e-21 | 0.7846153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200563 | CE4_HpPgdA_like | 1.11e-128 | 8 | 294 | 2 | 258 | Catalytic domain of Helicobacter pylori peptidoglycan deacetylase (HpPgdA) and similar proteins. This family is represented by a peptidoglycan deacetylase (HP0310, HpPgdA) from the gram-negative pathogen Helicobacter pylori. HpPgdA has the ability to bind a metal ion at the active site and is responsible for a peptidoglycan modification that counteracts the host immune response. It functions as a homotetramer. The monomer is composed of a 7-stranded barrel with detectable sequence similarity to the 6-stranded barrel NodB homology domain of polysaccharide deacetylase (DCA)-like proteins in the CE4 superfamily, which removes N-linked or O-linked acetyl groups from cell wall polysaccharides. In contrast to typical NodB-like DCAs, HpPgdA does not exhibit a solvent-accessible polysaccharide binding groove, suggesting that the enzyme binds a small molecule at the active site. |
| 213021 | CE4_PuuE_HpPgdA_like | 1.14e-60 | 8 | 288 | 2 | 243 | Catalytic domain of bacterial PuuE allantoinases, Helicobacter pylori peptidoglycan deacetylase (HpPgdA), and similar proteins. This family is a member of the very large and functionally diverse carbohydrate esterase 4 (CE4) superfamily. It contains bacterial PuuE (purine utilization E) allantoinases, a peptidoglycan deacetylase from Helicobacter pylori (HpPgdA), Escherichia coli ArnD, and many uncharacterized homologs from all three kingdoms of life. PuuE allantoinase appears to be metal-independent and specifically catalyzes the hydrolysis of (S)-allantoin into allantoic acid. Different from PuuE allantoinase, HpPgdA has the ability to bind a metal ion at the active site and is responsible for a peptidoglycan modification that counteracts the host immune response. Both PuuE allantoinase and HpPgdA function as a homotetramer. The monomer is composed of a 7-stranded barrel with detectable sequence similarity to the 6-stranded barrel NodB homology domain of polysaccharide deacetylase (DCA)-like proteins in the CE4 superfamily, which removes N-linked or O-linked acetyl groups from cell wall polysaccharides. However, in contrast with the typical DCAs, PuuE allantoinase and HpPgdA might not exhibit a solvent-accessible polysaccharide binding groove and only recognize a small substrate molecule. ArnD catalyzes the deformylation of 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol to 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol. |
| 200601 | CE4_PuuE_like | 6.72e-28 | 42 | 296 | 63 | 281 | Putative catalytic domain of uncharacterized prokaryotic polysaccharide deacetylases similar to bacterial PuuE allantoinases. The family includes a group of uncharacterized prokaryotic polysaccharide deacetylases (DCAs) that show high sequence similarity to the catalytic domain of bacterial PuuE (purine utilization E) allantoinases. PuuE allantoinase specifically catalyzes the hydrolysis of (S)-allantoin into allantoic acid. It functions as a homotetramer. Its monomer is composed of a 7-stranded barrel with detectable sequence similarity to the 6-stranded barrel NodB homology domain of DCA-like proteins in the CE4 superfamily, which removes N-linked or O-linked acetyl groups from cell wall polysaccharides. PuuE allantoinase appears to be metal-independent and acts on a small substrate molecule, which is distinct from the common feature of DCAs which are normally metal ion dependent and recognize multimeric substrates. |
| 213022 | CE4_NodB_like_6s_7s | 9.06e-23 | 44 | 185 | 16 | 147 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| 223798 | CDA1 | 2.08e-22 | 43 | 154 | 79 | 192 | Peptidoglycan/xylan/chitin deacetylase, PgdA/CDA1 family [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.39e-22 | 8 | 289 | 18 | 266 | |
| 1.21e-13 | 47 | 191 | 82 | 212 | |
| 1.27e-13 | 42 | 139 | 168 | 265 | |
| 5.86e-13 | 44 | 136 | 70 | 162 | |
| 8.22e-13 | 44 | 139 | 25 | 120 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.22e-141 | 4 | 297 | 36 | 322 | Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_B Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_C Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_D Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],4LY4_A Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_B Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_C Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_D Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27] |
|
| 5.23e-12 | 44 | 130 | 36 | 122 | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
|
| 1.81e-11 | 44 | 122 | 36 | 114 | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579],5O6Y_C Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579],5O6Y_D Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
|
| 3.59e-11 | 44 | 122 | 88 | 166 | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_B Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_C Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_D Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579] |
|
| 4.69e-08 | 46 | 113 | 103 | 170 | Crystal structure of a BA3943 mutant,a CE4 family pseudoenzyme [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-140 | 4 | 297 | 3 | 289 | Peptidoglycan deacetylase OS=Helicobacter pylori (strain G27) OX=563041 GN=pgdA PE=1 SV=1 |
|
| 2.84e-138 | 4 | 297 | 3 | 289 | Peptidoglycan deacetylase OS=Helicobacter pylori (strain ATCC 700392 / 26695) OX=85962 GN=pgdA PE=1 SV=1 |
|
| 1.38e-73 | 3 | 293 | 37 | 330 | Peptidoglycan deacetylase-like protein FGM2 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FGM2 PE=2 SV=1 |
|
| 5.61e-11 | 44 | 130 | 96 | 182 | Peptidoglycan-N-acetylglucosamine deacetylase BC_1960 OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=BC_1960 PE=1 SV=1 |
|
| 2.85e-08 | 44 | 124 | 82 | 169 | Uncharacterized 30.6 kDa protein in fumA 3'region OS=Geobacillus stearothermophilus OX=1422 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
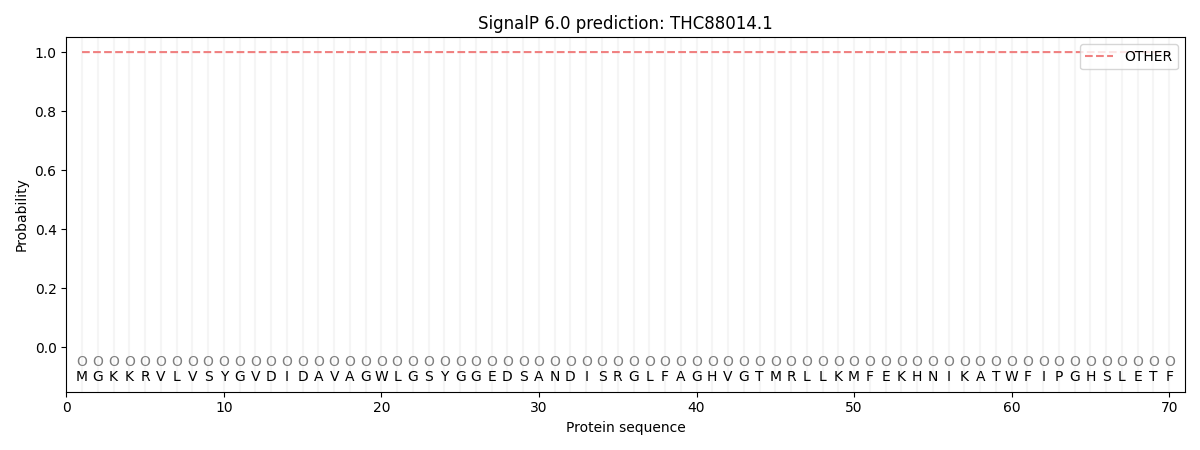
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000000 |
