You are browsing environment: FUNGIDB
CAZyme Information: TDH72551.1
You are here: Home > Sequence: TDH72551.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
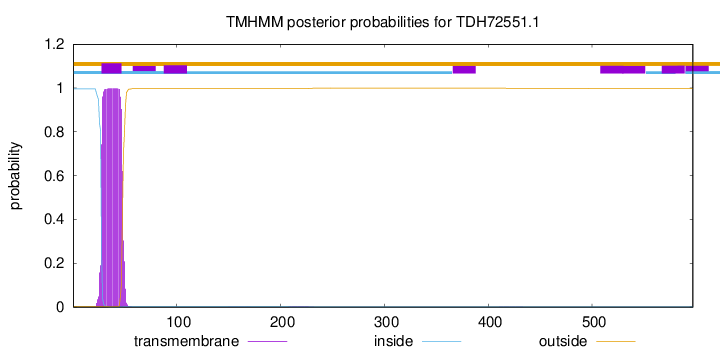
TMHMM annotations
Basic Information help
| Species | Bremia lactucae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Bremia; Bremia lactucae | |||||||||||
| CAZyme ID | TDH72551.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Glycosyltransferase family 71 protein [Source:UniProtKB/TrEMBL;Acc:A0A484E6G7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 208 | 442 | 1.9e-49 | 0.9924242424242424 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 1.23e-41 | 209 | 442 | 3 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.82e-91 | 212 | 585 | 1 | 360 | |
| 1.19e-90 | 104 | 488 | 106 | 516 | |
| 1.50e-90 | 76 | 478 | 71 | 475 | |
| 1.68e-87 | 4 | 582 | 630 | 1176 | |
| 4.10e-86 | 101 | 476 | 65 | 419 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-14 | 202 | 442 | 165 | 440 | Alpha-1,2-mannosyltransferase MNN2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN2 PE=1 SV=1 |
|
| 1.30e-13 | 208 | 462 | 150 | 453 | Alpha-1,2-mannosyltransferase MNN5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN5 PE=1 SV=2 |
|
| 3.00e-13 | 208 | 442 | 150 | 433 | Alpha-1,2-mannosyltransferase MNN5 OS=Saccharomyces cerevisiae (strain YJM789) OX=307796 GN=MNN5 PE=3 SV=1 |
|
| 9.69e-12 | 199 | 407 | 267 | 499 | Putative alpha-1,3-mannosyltransferase MNN14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN14 PE=2 SV=1 |
|
| 2.03e-11 | 208 | 440 | 174 | 447 | Alpha-1,2-mannosyltransferase MNN23 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN23 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000015 | 0.000014 |