You are browsing environment: FUNGIDB
CAZyme Information: TDH64793.1
You are here: Home > Sequence: TDH64793.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bremia lactucae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Bremia; Bremia lactucae | |||||||||||
| CAZyme ID | TDH64793.1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | EGF-like domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A484DQI2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH72 | 26 | 340 | 5.6e-73 | 0.8878205128205128 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397351 | Glyco_hydro_72 | 6.98e-39 | 18 | 378 | 1 | 315 | Glucanosyltransferase. This is a family of glycosylphosphatidylinositol-anchored beta(1-3)glucanosyltransferases. The active site residues in the Aspergillus fumigatus example are the two glutamate residues at 160 and 261. |
| 238011 | EGF_CA | 0.001 | 477 | 509 | 5 | 37 | Calcium-binding EGF-like domain, present in a large number of membrane-bound and extracellular (mostly animal) proteins. Many of these proteins require calcium for their biological function and calcium-binding sites have been found to be located at the N-terminus of particular EGF-like domains; calcium-binding may be crucial for numerous protein-protein interactions. Six conserved core cysteines form three disulfide bridges as in non calcium-binding EGF domains, whose structures are very similar. EGF_CA can be found in tandem repeat arrangements. |
| 238010 | EGF | 0.008 | 477 | 506 | 2 | 31 | Epidermal growth factor domain, found in epidermal growth factor (EGF) presents in a large number of proteins, mostly animal; the list of proteins currently known to contain one or more copies of an EGF-like pattern is large and varied; the functional significance of EGF-like domains in what appear to be unrelated proteins is not yet clear; a common feature is that these repeats are found in the extracellular domain of membrane-bound proteins or in proteins known to be secreted (exception: prostaglandin G/H synthase); the domain includes six cysteine residues which have been shown to be involved in disulfide bonds; the main structure is a two-stranded beta-sheet followed by a loop to a C-terminal short two-stranded sheet; Subdomains between the conserved cysteines vary in length; the region between the 5th and 6th cysteine contains two conserved glycines of which at least one is present in most EGF-like domains; a subset of these bind calcium. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 3 | 572 | 7 | 576 | |
| 1.90e-224 | 27 | 571 | 332 | 866 | |
| 2.03e-224 | 27 | 567 | 23 | 554 | |
| 9.34e-181 | 24 | 472 | 57 | 505 | |
| 6.77e-151 | 11 | 511 | 10 | 500 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-23 | 27 | 376 | 36 | 342 | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose [Saccharomyces cerevisiae],2W63_A Saccharomyces Cerevisiae Gas2p In Complex With Laminaritriose And Laminaritetraose [Saccharomyces cerevisiae],5O9O_A Crystal structure of ScGas2 in complex with compound 7. [Saccharomyces cerevisiae S288C],5O9P_A Crystal structure of Gas2 in complex with compound 10 [Saccharomyces cerevisiae S288C],5O9Q_A Crystal structure of ScGas2 in complex with compound 6 [Saccharomyces cerevisiae S288C],5O9R_A Crystal structure of ScGas2 in complex with compound 9 [Saccharomyces cerevisiae S288C],5O9Y_A Crystal structure of ScGas2 in complex with compound 11 [Saccharomyces cerevisiae S288C],5OA2_A Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA2_B Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA2_C Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA6_A Crystal structure of ScGas2 in complex with compound 12 [Saccharomyces cerevisiae S288C] |
|
| 2.99e-23 | 27 | 376 | 36 | 342 | Saccharomyces cerevisiae Gas2p apostructure (E176Q mutant) [Saccharomyces cerevisiae] |
|
| 2.99e-23 | 27 | 376 | 36 | 342 | SACCHAROMYCES CEREVISIAE GAS2P (E176Q MUTANT) IN COMPLEX WITH LAMINARITETRAOSE AND LAMINARIPENTAOSE [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.34e-27 | 27 | 397 | 22 | 341 | 1,3-beta-glucanosyltransferase PGA4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PGA4 PE=1 SV=1 |
|
| 3.83e-25 | 13 | 408 | 16 | 362 | 1,3-beta-glucanosyltransferase GAS5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS5 PE=1 SV=1 |
|
| 6.49e-23 | 27 | 376 | 36 | 342 | 1,3-beta-glucanosyltransferase GAS2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS2 PE=1 SV=1 |
|
| 1.21e-22 | 18 | 389 | 20 | 347 | 1,3-beta-glucanosyltransferase gas5 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gas5 PE=1 SV=1 |
|
| 2.20e-22 | 27 | 374 | 26 | 331 | 1,3-beta-glucanosyltransferase GAS4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
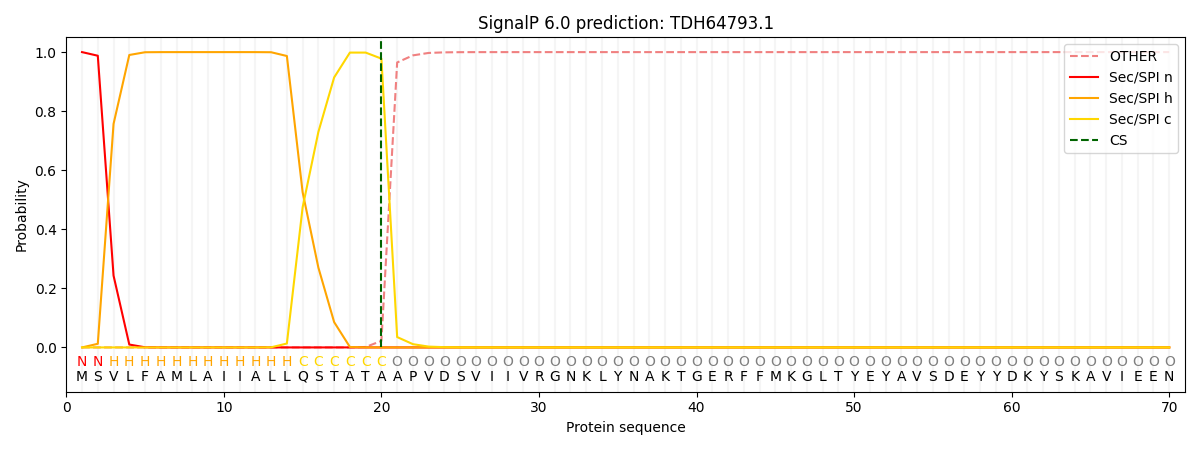
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000192 | 0.999777 | CS pos: 20-21. Pr: 0.9780 |

