You are browsing environment: FUNGIDB
CAZyme Information: SPSK_08401-t39_1-p1
You are here: Home > Sequence: SPSK_08401-t39_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
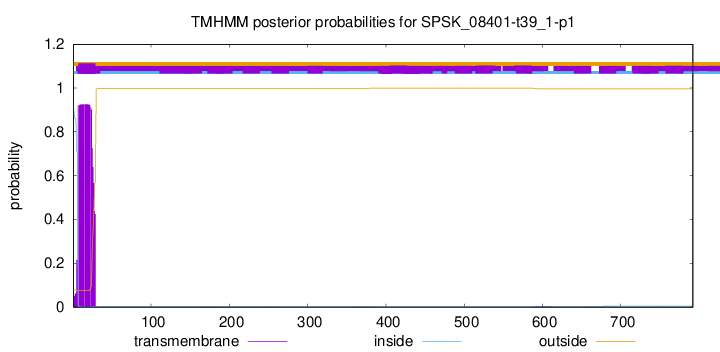
TMHMM annotations
Basic Information help
| Species | Sporothrix schenckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix schenckii | |||||||||||
| CAZyme ID | SPSK_08401-t39_1-p1 | |||||||||||
| CAZy Family | GH79 | |||||||||||
| CAZyme Description | glycoside hydrolase family 55 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403800 | Pectate_lyase_3 | 4.81e-62 | 197 | 443 | 1 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 403800 | Pectate_lyase_3 | 1.37e-04 | 570 | 630 | 10 | 74 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.25e-296 | 22 | 792 | 13 | 721 | |
| 5.71e-279 | 40 | 791 | 36 | 728 | |
| 1.31e-278 | 49 | 791 | 1 | 708 | |
| 1.95e-276 | 49 | 792 | 1 | 680 | |
| 2.55e-272 | 49 | 791 | 1 | 704 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.46e-140 | 172 | 790 | 2 | 611 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
|
| 8.09e-132 | 169 | 791 | 19 | 625 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.72e-123 | 165 | 791 | 32 | 738 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
|
| 1.07e-105 | 170 | 790 | 42 | 645 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 7.18e-34 | 204 | 735 | 64 | 564 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.270151 | 0.729839 | CS pos: 24-25. Pr: 0.6799 |