You are browsing environment: FUNGIDB
CAZyme Information: SPSK_00893-t39_1-p1
You are here: Home > Sequence: SPSK_00893-t39_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporothrix schenckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix schenckii | |||||||||||
| CAZyme ID | SPSK_00893-t39_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:59 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 28 | 230 | 5.1e-91 | 0.9949494949494949 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 5.45e-78 | 27 | 230 | 1 | 214 | Glycosyl hydrolase family 45. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.20e-131 | 6 | 231 | 7 | 227 | |
| 3.61e-128 | 7 | 231 | 3 | 229 | |
| 9.94e-128 | 14 | 231 | 10 | 227 | |
| 2.03e-126 | 15 | 231 | 14 | 233 | |
| 2.98e-126 | 16 | 231 | 15 | 225 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-98 | 14 | 232 | 13 | 224 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 1.61e-98 | 24 | 232 | 1 | 203 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
|
| 1.85e-98 | 24 | 232 | 4 | 206 | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126] |
|
| 6.20e-96 | 28 | 232 | 3 | 202 | Endoglucanase from Humicola insolens AT 1.7A resolution [Humicola insolens] |
|
| 2.05e-95 | 26 | 232 | 1 | 201 | Structure of 20K-endoglucanase from Melanocarpus albomyces at 1.8 A [Melanocarpus albomyces] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.61e-94 | 28 | 232 | 3 | 202 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
|
| 9.13e-93 | 24 | 232 | 18 | 222 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 1.26e-85 | 10 | 231 | 5 | 221 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 1.80e-56 | 23 | 229 | 35 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 1.69e-48 | 28 | 232 | 269 | 501 | Endoglucanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
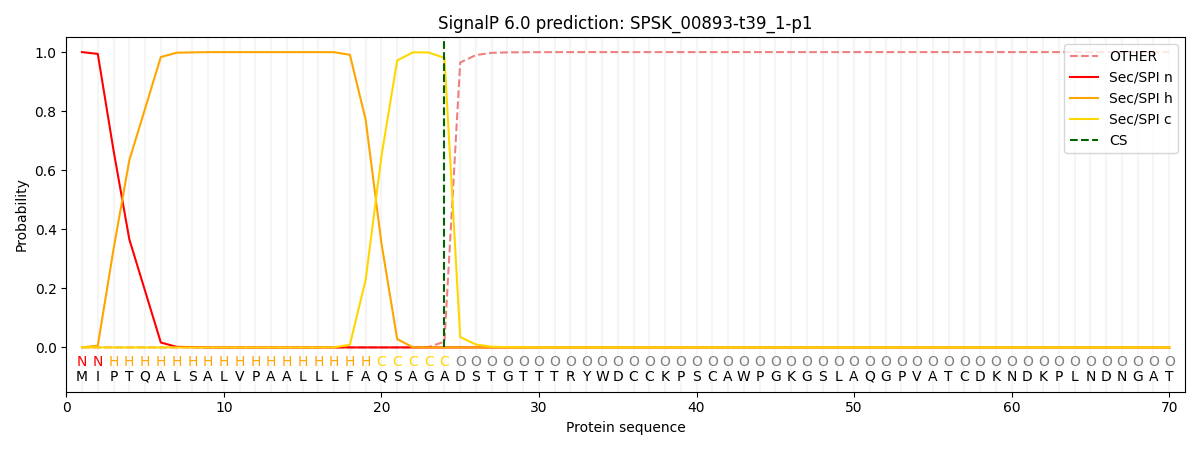
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000213 | 0.999763 | CS pos: 24-25. Pr: 0.9804 |
