You are browsing environment: FUNGIDB
CAZyme Information: SPRG_22219-t26_1-p1
You are here: Home > Sequence: SPRG_22219-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_22219-t26_1-p1 | |||||||||||
| CAZy Family | GT61 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 197844; End:201385 Strand: + | |||||||||||
Full Sequence Download help
| MQATSAWSGH AVVAALAAAT FANTLTCGFV WDDRAAILTN RDLRTDETSM WDLWSHDFGA | 60 |
| GHSLLTFRAN YAVGGYNPMG YHLLNVLLHV AASVLVVAVG RRVVAPFGAS HAPVFAGALF | 120 |
| AVHPIHCDAV ASVVGRADVL CTCLSLLAFC LYDASASATC THWPLYVASL VCILAATLCK | 180 |
| ELGAATLGIL VTLELLRAAP SKSKRLGPSA GVVRTAVLFG LGSAGIASRI ALNGGQTLYA | 240 |
| WSILENDVSL LPWGVPRLLT TLHTHGWYLY KLFWPQYLCN DYGFRTIPII ASLFQMENVG | 300 |
| TLVAYASLLV LVGVAFDRRR TSPLLLLASF GLFPFVPAAN VLFPIGTVVA ERLMYFPSVG | 360 |
| FCFVLGYALE HALRVSMRWQ SLLLALLFLS LLTVGAARSM ARNFEWSSET RLFESSVLTA | 420 |
| PWSVKGLSNL SKVLLGPDPP RAAAYLERAL SVAPRYAAGQ LNLGLAYIRM GKPLHAMQNL | 480 |
| LHSIDVDASL AAYAYLGRCA SSFYLASQLK QFPTSNATHA CQLAHQLLDY TIAQGASMPG | 540 |
| LFFARGLLAY YTSDFSTAFA VCANQLTTPM NQQQPTTTAY EIQPTPTVKP VAPDGMEADA | 600 |
| NNLVVGQWKV GICGCFTDVI PNCCMAYWCP CVSLAQIVHR IGTCSYMMGL LVFGLAYLLM | 660 |
| YITAAVSGGY HYSAIASGSA GVACGICVMI VRNIVRGKLR IPGNCLIDCL CGFFCNCCAI | 720 |
| AQMATQVNAY DKGTCAFGPK DTLPGYMA | 748 |
Enzyme Prediction help
| EC | 2.4.1.109:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT105 | 70 | 199 | 6e-33 | 0.9236641221374046 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398426 | PLAC8 | 1.78e-20 | 608 | 724 | 1 | 98 | PLAC8 family. This family includes the Placenta-specific gene 8 protein. |
| 273697 | A_thal_Cys_rich | 6.80e-12 | 606 | 728 | 1 | 103 | uncharacterized Cys-rich domain. This model describes an uncharacterized domain of about 100 residues. It is common in plants but found also in Homo sapiens, Dictyostelium, and Leishmania; at least 12 distinct members are found in Arabidopsis. Most members of this family contain more than 10 per cent Cys, but no Cys residue is invariant across the family. |
| 276809 | TPR | 9.31e-06 | 438 | 499 | 16 | 78 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 1.54e-04 | 411 | 485 | 22 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 274350 | PEP_TPR_lipo | 5.95e-04 | 396 | 495 | 777 | 875 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAD7262601.1|GT105 | 1.32e-50 | 19 | 467 | 17 | 490 |
| Q5T4D3|GT105|2.4.1.109 | 6.85e-43 | 12 | 443 | 25 | 501 |
| BAF83034.1|GT105|2.4.1.109 | 9.22e-43 | 12 | 443 | 25 | 501 |
| AAI66274.1|GT105 | 2.96e-42 | 12 | 456 | 24 | 511 |
| BAC25886.1|GT105 | 8.79e-42 | 21 | 443 | 33 | 501 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q5T4D3|TMTC4_HUMAN | 1.22e-43 | 12 | 443 | 25 | 501 | Protein O-mannosyl-transferase TMTC4 OS=Homo sapiens OX=9606 GN=TMTC4 PE=1 SV=2 |
| sp|Q8BG19|TMTC4_MOUSE | 5.77e-42 | 21 | 443 | 33 | 501 | Protein O-mannosyl-transferase TMTC4 OS=Mus musculus OX=10090 GN=Tmtc4 PE=2 SV=1 |
| sp|Q6ZXV5|TMTC3_HUMAN | 7.33e-42 | 12 | 479 | 11 | 502 | Protein O-mannosyl-transferase TMTC3 OS=Homo sapiens OX=9606 GN=TMTC3 PE=1 SV=2 |
| sp|Q8BRH0|TMTC3_MOUSE | 1.32e-41 | 12 | 476 | 16 | 504 | Protein O-mannosyl-transferase TMTC3 OS=Mus musculus OX=10090 GN=Tmtc3 PE=1 SV=2 |
| sp|Q7K4B6|TMTC3_DROME | 5.69e-41 | 23 | 478 | 50 | 569 | Protein O-mannosyl-transferase Tmtc3 OS=Drosophila melanogaster OX=7227 GN=Tmtc3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.776122 | 0.223876 |
TMHMM Annotations download full data without filtering help
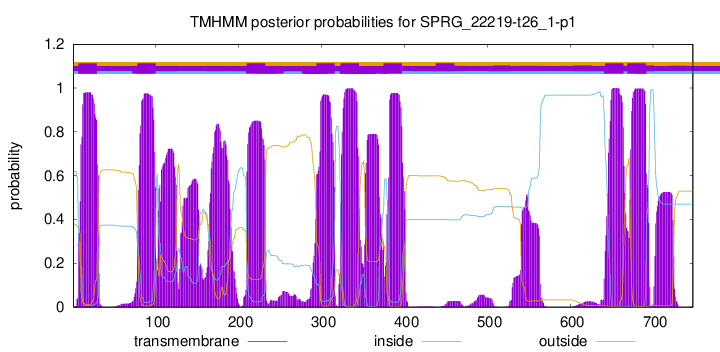
| Start | End |
|---|---|
| 7 | 29 |
| 78 | 100 |
| 210 | 232 |
| 294 | 316 |
| 323 | 345 |
| 375 | 397 |
| 642 | 664 |
| 669 | 691 |
