You are browsing environment: FUNGIDB
CAZyme Information: SPRG_22161-t26_1-p1
You are here: Home > Sequence: SPRG_22161-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_22161-t26_1-p1 | |||||||||||
| CAZy Family | GT60 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 8 | 233 | 5e-73 | 0.9224489795918367 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395595 | CBM_1 | 3.33e-12 | 239 | 266 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 3.95e-11 | 237 | 270 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 223880 | TonB | 0.003 | 191 | 233 | 61 | 102 | Periplasmic protein TonB, links inner and outer membranes [Cell wall/membrane/envelope biogenesis]. |
| 411470 | opacity_OapA | 0.008 | 190 | 238 | 222 | 270 | opacity-associated protein OapA. This family consists of full-length homologs to OapA, opacity-associated protein A as described in Haemophilus influenzae. OapA shares a C-terminal homology domain, called the OapA domain, with the Escherichia coli protein YtfB, which is now known to bind peptidoglycan through its OapA domain and to act as a cell division protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.17e-215 | 1 | 300 | 1 | 300 | |
| 1.02e-144 | 1 | 277 | 1 | 254 | |
| 8.22e-139 | 1 | 271 | 1 | 254 | |
| 1.15e-128 | 1 | 271 | 1 | 234 | |
| 2.32e-128 | 1 | 271 | 1 | 234 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.87e-32 | 21 | 188 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
|
| 4.88e-08 | 239 | 269 | 7 | 37 | Chain A, ENDOGLUCANASE EG-1 [Trichoderma reesei] |
|
| 1.10e-06 | 235 | 269 | 1 | 35 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 2.07e-06 | 235 | 269 | 1 | 35 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 5.31e-06 | 240 | 269 | 6 | 35 | Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],1AZH_A Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],5X3C_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.12e-09 | 237 | 275 | 19 | 57 | Endo-1,4-beta-xylanase D OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLD PE=1 SV=1 |
|
| 4.16e-09 | 238 | 277 | 21 | 60 | Putative endoglucanase type F OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 7.00e-08 | 222 | 270 | 466 | 514 | Putative exoglucanase type C OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 1.60e-07 | 239 | 276 | 30 | 67 | Exoglucanase-6A OS=Humicola insolens OX=34413 GN=cel6A PE=1 SV=1 |
|
| 2.09e-07 | 238 | 273 | 22 | 57 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
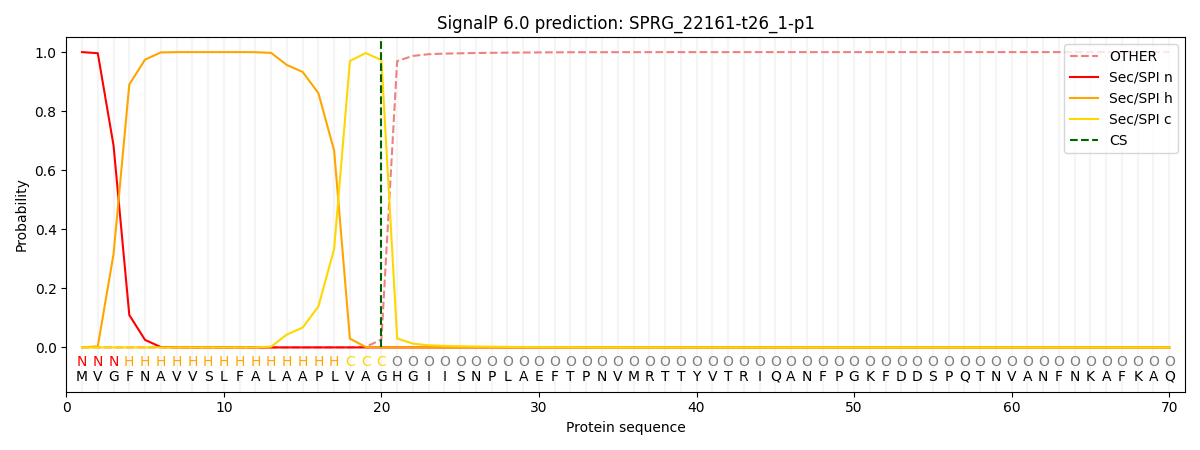
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000273 | 0.999705 | CS pos: 20-21. Pr: 0.9733 |

