You are browsing environment: FUNGIDB
CAZyme Information: SPRG_20839-t26_1-p1
You are here: Home > Sequence: SPRG_20839-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_20839-t26_1-p1 | |||||||||||
| CAZy Family | GT60 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.50:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 292 | 963 | 4.4e-219 | 0.9939668174962293 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398659 | NAGLU | 1.53e-179 | 351 | 686 | 2 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| 404009 | NAGLU_C | 1.34e-81 | 694 | 960 | 1 | 258 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| 340874 | MFS_SV2_like | 7.87e-37 | 33 | 209 | 4 | 179 | Metazoan Synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters of the Major Facilitator Superfamily. This family is composed of metazoan synaptic vesicle glycoprotein 2 (SV2) and related small molecule transporters including those that transport inorganic phosphate (Pht), aromatic compounds (PcaK and related proteins), proline/betaine (ProP), alpha-ketoglutarate (KgtP), citrate (CitA), shikimate (ShiA), and cis,cis-muconate (MucK), among others. SV2 is a transporter-like protein that serves as the receptor for botulinum neurotoxin A (BoNT/A), one of seven neurotoxins produced by the bacterium Clostridium botulinum. BoNT/A blocks neurotransmitter release by cleaving synaptosome-associated protein of 25 kD (SNAP-25) within presynaptic nerve terminals. Also included in this family is synaptic vesicle 2 (SV2)-related protein (SVOP) and similar proteins. SVOP is a transporter-like nucleotide binding protein that localizes to neurotransmitter-containing vesicles. The SV2-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340929 | MFS_MucK | 6.29e-28 | 35 | 219 | 6 | 187 | Cis,cis-muconate transport protein and similar proteins of the Major Facilitator Superfamily. This subfamily is composed of Acinetobacter sp. Cis,cis-muconate transport protein (MucK), Escherichia coli putative sialic acid transporter 1, and similar proteins. MucK functions in the uptake of muconate and allows Acinetobacter calcoaceticus ADP1 (BD413) to grow on exogenous cis,cis-muconate as the sole carbon source. The MucK subfamily belongs to the Metazoan Synaptic Vesicle Glycoprotein 2 (SV2) and related small molecule transporter family (SV2-like) of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 395036 | Sugar_tr | 5.86e-27 | 30 | 215 | 7 | 197 | Sugar (and other) transporter. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 231 | 982 | 8 | 754 | |
| 0.0 | 242 | 981 | 15 | 744 | |
| 1.88e-284 | 230 | 969 | 12 | 751 | |
| 2.28e-273 | 232 | 966 | 13 | 747 | |
| 6.94e-262 | 231 | 966 | 10 | 748 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.38e-162 | 248 | 967 | 2 | 712 | Crystal structure of the human N-acetyl-alpha-glucosaminidase [Homo sapiens] |
|
| 5.19e-91 | 243 | 912 | 162 | 822 | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
|
| 6.01e-91 | 243 | 912 | 170 | 830 | Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
|
| 5.41e-90 | 243 | 912 | 185 | 845 | CpGH89 (E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
|
| 2.69e-07 | 4 | 214 | 27 | 228 | Chain A, Sugar transport protein 10 [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.42e-161 | 221 | 967 | 1 | 735 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
|
| 5.41e-155 | 294 | 967 | 94 | 805 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
|
| 1.04e-28 | 29 | 222 | 12 | 198 | Putative niacin/nicotinamide transporter NaiP OS=Bacillus subtilis (strain 168) OX=224308 GN=naiP PE=1 SV=1 |
|
| 8.62e-15 | 35 | 222 | 40 | 220 | Sialic acid transporter NanT OS=Yersinia pseudotuberculosis serotype O:3 (strain YPIII) OX=502800 GN=nanT PE=3 SV=1 |
|
| 1.51e-14 | 35 | 217 | 22 | 197 | Putative metabolite transport protein YjhB OS=Escherichia coli (strain K12) OX=83333 GN=yjhB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.909102 | 0.090888 |
TMHMM Annotations download full data without filtering help
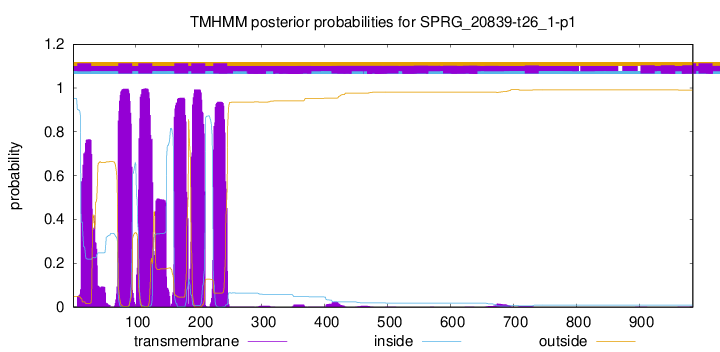
| Start | End |
|---|---|
| 72 | 94 |
| 104 | 126 |
| 161 | 183 |
| 188 | 210 |
| 223 | 245 |
