You are browsing environment: FUNGIDB
CAZyme Information: SPRG_20259-t26_1-p1
You are here: Home > Sequence: SPRG_20259-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_20259-t26_1-p1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | - | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 1566 | 2213 | 1.6e-260 | 0.9242219215155616 |
| GH16 | 62 | 399 | 4.4e-90 | 0.9969325153374233 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 1566 | 2196 | 3 | 712 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 397841 | SKN1 | 1.63e-86 | 25 | 442 | 96 | 489 | Beta-glucan synthesis-associated protein (SKN1). This family consists of the beta-glucan synthesis-associated proteins KRE6 and SKN1. Beta1,6-Glucan is a key component of the yeast cell wall, interconnecting cell wall proteins, beta1,3-glucan, and chitin. It has been postulated that the synthesis of beta1,6-glucan begins in the endoplasmic reticulum with the formation of protein-bound primer structures and that these primer structures are extended in the Golgi complex by two putative glucosyltransferases that are functionally redundant, Kre6 and Skn1. This is followed by maturation steps at the cell surface and by coupling to other cell wall macromolecules. |
| 185689 | GH16_fungal_KRE6_glucanase | 7.22e-86 | 58 | 400 | 1 | 295 | Saccharomyces cerevisiae KRE6 and related glucanses, member of glycosyl hydrolase family 16. KRE6 is a Saccharomyces cerevisiae glucanase that participates in the synthesis of beta-1,6-glucan, a major structural component of the cell wall. It is a golgi membrane protein required for normal beta-1,6-glucan levels in the cell wall. KRE6 is closely realted to laminarinase, a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 185693 | GH16_laminarinase_like | 1.58e-30 | 60 | 399 | 1 | 235 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 405046 | FKS1_dom1 | 4.22e-28 | 765 | 856 | 1 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 26 | 2428 | 38 | 2522 | |
| 0.0 | 21 | 2433 | 25 | 2513 | |
| 6.08e-228 | 705 | 2389 | 45 | 1715 | |
| 3.89e-226 | 705 | 2389 | 247 | 1917 | |
| 8.76e-225 | 705 | 2384 | 247 | 1945 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-08 | 58 | 400 | 13 | 256 | Crystal structure of of LamAcat from Zobellia galactanivorans [Zobellia galactanivorans],4BQ1_B Crystal structure of of LamAcat from Zobellia galactanivorans [Zobellia galactanivorans] |
|
| 3.28e-08 | 58 | 400 | 13 | 256 | Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose [Zobellia galactanivorans],4BOW_B Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose [Zobellia galactanivorans],4BPZ_A Crystal structure of lamA_E269S from Zobellia galactanivorans in complex with a trisaccharide of 1,3-1,4-beta-D-glucan. [Zobellia galactanivorans],4BPZ_B Crystal structure of lamA_E269S from Zobellia galactanivorans in complex with a trisaccharide of 1,3-1,4-beta-D-glucan. [Zobellia galactanivorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.25e-220 | 705 | 2384 | 241 | 1937 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
|
| 1.54e-217 | 705 | 2384 | 245 | 1942 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 2.71e-216 | 705 | 2389 | 248 | 1921 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 1.02e-215 | 701 | 2384 | 260 | 1921 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 3.98e-214 | 705 | 2384 | 241 | 1937 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.022509 | 0.977474 | CS pos: 19-20. Pr: 0.8743 |
TMHMM Annotations download full data without filtering help
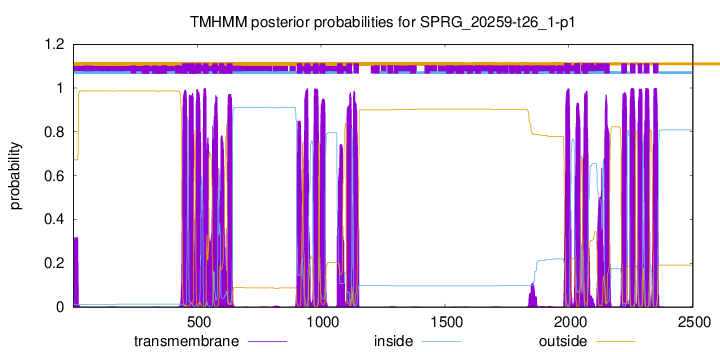
| Start | End |
|---|---|
| 439 | 461 |
| 468 | 490 |
| 495 | 517 |
| 524 | 546 |
| 561 | 583 |
| 588 | 610 |
| 620 | 642 |
| 902 | 924 |
| 934 | 956 |
| 968 | 990 |
| 994 | 1016 |
| 1065 | 1087 |
| 1102 | 1124 |
| 1131 | 1153 |
| 1982 | 2004 |
| 2024 | 2046 |
| 2056 | 2078 |
| 2143 | 2165 |
| 2212 | 2234 |
| 2247 | 2269 |
| 2279 | 2296 |
| 2303 | 2325 |
| 2340 | 2362 |
