You are browsing environment: FUNGIDB
CAZyme Information: SPRG_19298-t26_1-p1
You are here: Home > Sequence: SPRG_19298-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_19298-t26_1-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 523079; End:527544 Strand: - | |||||||||||
Full Sequence Download help
| MADTLASAFA LMQAGARGEA QERLIALARI APQNADVHTA LGALAQMDGH VDRAIASYAT | 60 |
| ALSLCGPTEA LHGNLGLAHY ARQDYKASVE HFRAAIALNP ARLPDLAHML GLALHFLRDD | 120 |
| AAAKDMYVAA VAHAPHDAAV RFDYGVTLQA LGDIEQAGDA YNRAIALNPA MGSAWLNMAS | 180 |
| LHLQYGEVNK ALRGFEKTLG LPLPIDLWLC ATTNYAVALE LDGQPLAATK FLKRAHAVLQ | 240 |
| LKGATTSTLY LHVCEHQIRT WRAIAYWKDY ELVWTRFFEM TWQHEIQVGA VSSMMPFTSL | 300 |
| LLPLAPEMKR KIAESITRPH VSAEKRLWRA TPPVAGARRL HVGYLSYDFN NHPTAHLMEG | 360 |
| LFRCHNASSV EVSMLSYGKD DNSSYRRLFP TLVEHFVDLA RAGTRAAASV IRDAHVDILI | 420 |
| DAQGHTLGQR HDIVAQQPAP IIINYLVFPG TLGAPYVDYL LADVHVAPPE HAHHFVEKLL | 480 |
| YVPHSYQVNY FASPVPFSET RRTGRFVFAN YNKIDKLEPR VFSVWMQILR RVPRSELWLL | 540 |
| APTSTKTEQL TMRHVHMEAA VYGIPPSRIR FLPRVTKAAH LARQADADLF LDTFVYGAHS | 600 |
| TATDAMWGHL PVLTLAGDSF TSRVGISLAT NANSVELVVH SAKEFADVAD KDWIYDRAMS | 660 |
| SAAEVFTIMA AAVADAGEAL VKKVNGSIKF DVKGAGMWLI NLKAAPGAVT ASNAGEKADL | 720 |
| TITISEPDFV DLINEKLNPQ AAFMKGKIKV KGNMGLAMKL SAVTNATKAY LAKQKKSPAA | 780 |
| AAPVAAAPAA TSGLKSAALF VGIGEAVKTQ GPALVAKVKG TIQFNIAPGG AWFLDLKNGN | 840 |
| GSLETGSKPA DLTINVSDED FMAIADGKLN AQQAFMKGKL KVKGNMGLAM KLNIVIDAAK | 900 |
| PKAKL | 905 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 121 | 649 | 2.2e-92 | 0.5134751773049645 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 5.71e-74 | 148 | 645 | 46 | 567 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 404688 | Glyco_transf_41 | 4.16e-54 | 267 | 651 | 4 | 509 | Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. |
| 396566 | SCP2 | 1.50e-22 | 812 | 892 | 12 | 95 | SCP-2 sterol transfer family. This domain is involved in binding sterols. It is found in the SCP2 protein, as well as the C-terminus of the enzyme estradiol 17 beta-dehydrogenase EC:1.1.1.62. The UNC-24 protein contains an SPFH domain pfam01145. |
| 396566 | SCP2 | 1.92e-18 | 680 | 763 | 14 | 98 | SCP-2 sterol transfer family. This domain is involved in binding sterols. It is found in the SCP2 protein, as well as the C-terminus of the enzyme estradiol 17 beta-dehydrogenase EC:1.1.1.62. The UNC-24 protein contains an SPFH domain pfam01145. |
| 276809 | TPR | 3.57e-13 | 71 | 166 | 3 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQN61861.1|GT41 | 4.58e-75 | 70 | 649 | 54 | 611 |
| ALF56607.1|GT41 | 4.05e-74 | 339 | 649 | 16 | 327 |
| ALF56465.1|GT41 | 5.78e-74 | 339 | 649 | 23 | 334 |
| QCJ91786.1|GT41 | 8.14e-72 | 70 | 649 | 54 | 611 |
| QCJ84417.1|GT41 | 8.14e-72 | 70 | 649 | 54 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5A01_A | 2.94e-42 | 267 | 649 | 159 | 651 | O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_B O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_C O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster] |
| 2JLB_A | 2.05e-41 | 129 | 628 | 14 | 495 | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
| 2VSN_A | 3.71e-41 | 129 | 628 | 14 | 495 | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
| 6Q4M_A | 1.06e-40 | 267 | 651 | 161 | 664 | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation [Homo sapiens] |
| 5NPR_A | 1.35e-40 | 267 | 651 | 155 | 658 | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9M8Y0|SEC_ARATH | 2.18e-61 | 258 | 649 | 502 | 908 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
| sp|Q8CGY8|OGT1_MOUSE | 8.17e-40 | 267 | 651 | 479 | 982 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Mus musculus OX=10090 GN=Ogt PE=1 SV=2 |
| sp|Q27HV0|OGT1_PIG | 1.08e-39 | 267 | 651 | 479 | 982 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Sus scrofa OX=9823 GN=OGT PE=2 SV=1 |
| sp|P81436|OGT1_RABIT | 2.50e-39 | 267 | 651 | 479 | 982 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Oryctolagus cuniculus OX=9986 GN=OGT PE=1 SV=2 |
| sp|O15294|OGT1_HUMAN | 2.50e-39 | 267 | 651 | 479 | 982 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Homo sapiens OX=9606 GN=OGT PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
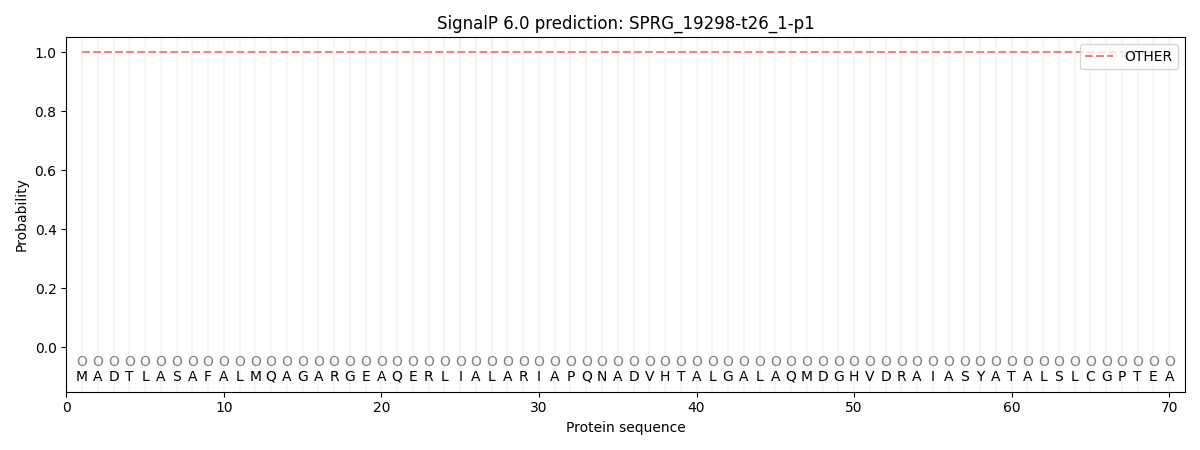
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000002 |
