You are browsing environment: FUNGIDB
CAZyme Information: SPRG_11812-t26_1-p1
You are here: Home > Sequence: SPRG_11812-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_11812-t26_1-p1 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 | 3.2.1.55:25 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH62 | 245 | 528 | 1.7e-114 | 0.9928057553956835 |
| GH11 | 29 | 209 | 1.1e-69 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350101 | GH62 | 2.16e-170 | 246 | 557 | 1 | 304 | Glycosyl hydrolase family 62, characterized arabinofuranosidases. The glycosyl hydrolase family 62 (GH62) includes eukaryotic (mostly fungal) and prokaryotic enzymes which are characterized arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. These enzymes show significantly different substrate preference with rather low specific activity towards natural substrates and differ in catalytic efficiency. They do not act on xylose moieties in xylan that are adorned with an arabinose side chain at both O2 and O3 positions, nor do they display any non-specific arabinofuranosidase activity. The synergistic action in biomass degradation makes GH62 promising candidates for biotechnological improvements of biofuel production and in various biorefinery applications. These enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. |
| 395367 | Glyco_hydro_11 | 2.86e-103 | 29 | 208 | 1 | 175 | Glycosyl hydrolases family 11. |
| 281639 | Glyco_hydro_62 | 5.10e-66 | 246 | 527 | 3 | 271 | Glycosyl hydrolase family 62. Family of alpha -L-arabinofuranosidase (EC 3.2.1.55). This enzyme hydrolyzed aryl alpha-L-arabinofuranosides and cleaves arabinosyl side chains from arabinoxylan and arabinan. |
| 350092 | GH_F | 3.10e-14 | 272 | 519 | 1 | 242 | Glycosyl hydrolase families 43 and 62 form CAZY clan GH-F. This glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) includes family 43 (GH43) and 62 (GH62). GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH62 includes enzymes characterized as arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. GH62 are also predicted to be inverting enzymes. A common structural feature of both, GH43 and GH62 enzymes, is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 282710 | PT | 5.34e-05 | 217 | 242 | 3 | 28 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.32e-310 | 8 | 563 | 7 | 575 | |
| 6.10e-166 | 244 | 559 | 16 | 329 | |
| 8.42e-166 | 244 | 560 | 25 | 339 | |
| 6.17e-156 | 242 | 560 | 55 | 370 | |
| 3.86e-155 | 245 | 560 | 32 | 345 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.14e-155 | 246 | 560 | 1 | 314 | Chain A, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6S_B Chain B, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6T_A Chain A, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6T_B Chain B, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130] |
|
| 3.06e-146 | 244 | 559 | 20 | 334 | Chain A, GH62 arabinofuranosidase [Podospora anserina],4N4B_A Chain A, GH62 arabinofuranosidase [Podospora anserina] |
|
| 4.71e-138 | 248 | 557 | 22 | 342 | Crystal structure of GH62 hydrolase in complex with xylotriose [Mycothermus thermophilus] |
|
| 2.16e-136 | 248 | 557 | 22 | 342 | Crystal structure of GH62 hydrolase from thermophilic fungus Scytalidium thermophilum [Mycothermus thermophilus] |
|
| 1.73e-77 | 21 | 211 | 7 | 192 | Crystal Structure of a Xylanase at 1.56 Angstroem resolution [Fusarium oxysporum],5JRN_A Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution [Fusarium oxysporum f. sp. vasinfectum 25433] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.75e-90 | 20 | 211 | 38 | 224 | Endo-1,4-beta-xylanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnA PE=1 SV=1 |
|
| 5.13e-87 | 19 | 211 | 40 | 227 | Endo-1,4-beta-xylanase xynf11a OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnA PE=1 SV=1 |
|
| 5.13e-87 | 19 | 211 | 40 | 227 | Probable endo-1,4-beta-xylanase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xlnA PE=3 SV=1 |
|
| 1.03e-85 | 20 | 211 | 38 | 224 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 5.72e-85 | 20 | 211 | 38 | 224 | Endo-1,4-beta-xylanase B OS=Aspergillus niger OX=5061 GN=xlnB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
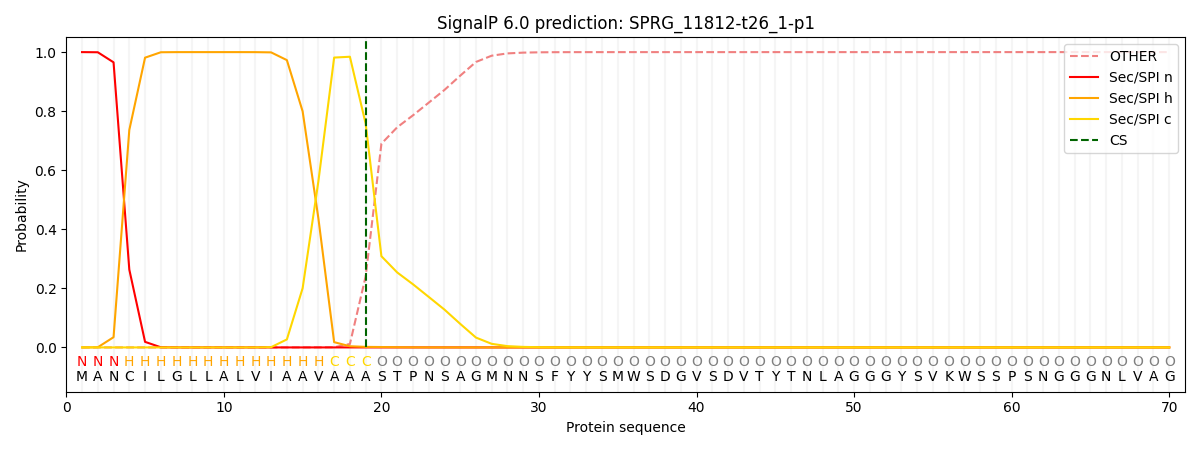
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000197 | 0.999763 | CS pos: 19-20. Pr: 0.7578 |

