You are browsing environment: FUNGIDB
CAZyme Information: SPRG_10264-t26_1-p1
Basic Information
help
| Species |
Saprolegnia parasitica
|
| Lineage |
Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica
|
| CAZyme ID |
SPRG_10264-t26_1-p1
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 821 |
KK583239|CGC1 |
87652.69 |
7.9358 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_SparasiticaCBS223-65 |
20435 |
695850 |
314 |
20121
|
|
| Gene Location |
No EC number prediction in SPRG_10264-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
206 |
592 |
3.8e-20 |
0.7001897533206831 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 404168
|
Beta_helix |
1.77e-05 |
473 |
596 |
25 |
126 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.40e-10 |
444 |
589 |
462 |
604 |
| 3.08e-10 |
367 |
599 |
251 |
495 |
| 4.29e-10 |
439 |
589 |
499 |
646 |
| 2.42e-09 |
367 |
616 |
267 |
508 |
| 3.16e-09 |
207 |
603 |
115 |
480 |
SPRG_10264-t26_1-p1 has no PDB hit.
SPRG_10264-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
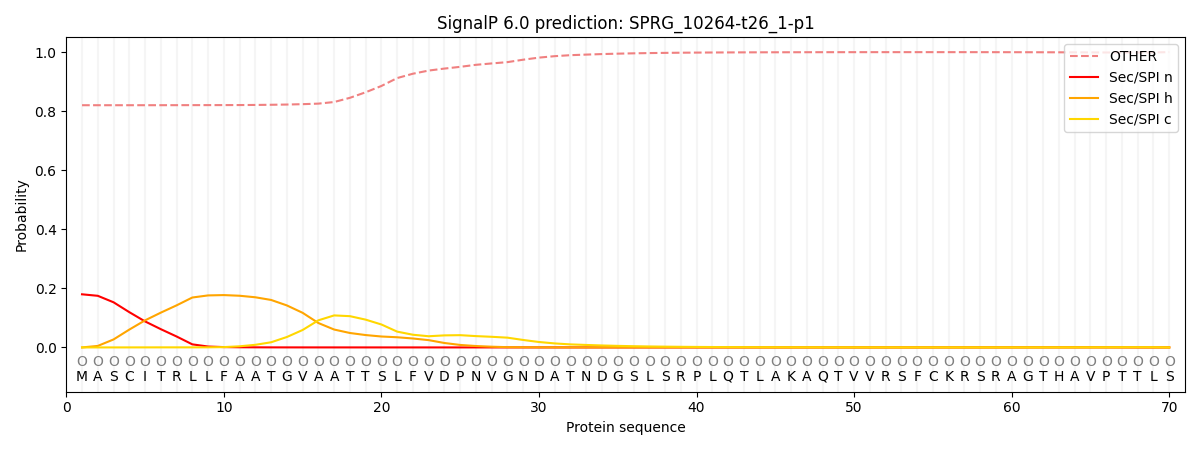
| Other |
SP_Sec_SPI |
CS Position |
| 0.832674 |
0.167328 |
|
There is no transmembrane helices in SPRG_10264-t26_1-p1.

