You are browsing environment: FUNGIDB
CAZyme Information: SPRG_10168-t26_1-p1
You are here: Home > Sequence: SPRG_10168-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
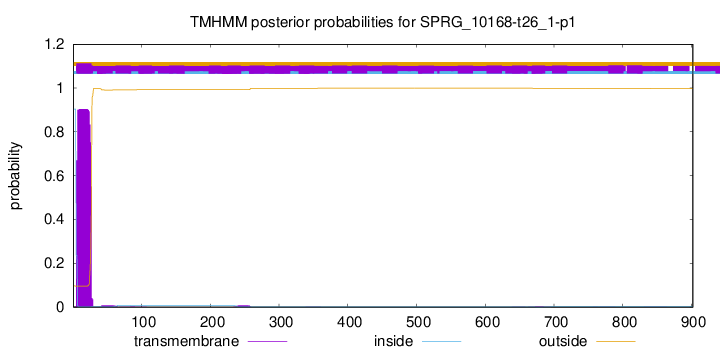
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_10168-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 374 | 585 | 7.2e-18 | 0.77491961414791 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.20e-15 | 294 | 580 | 32 | 303 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 227625 | Scw11 | 2.99e-11 | 604 | 797 | 21 | 206 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 227625 | Scw11 | 1.62e-10 | 52 | 278 | 67 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 8.36e-06 | 332 | 585 | 29 | 304 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 7 | 899 | 7 | 906 | |
| 4.80e-248 | 14 | 553 | 12 | 559 | |
| 9.33e-117 | 295 | 585 | 16 | 308 | |
| 4.57e-108 | 295 | 590 | 15 | 316 | |
| 4.12e-101 | 288 | 590 | 7 | 309 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
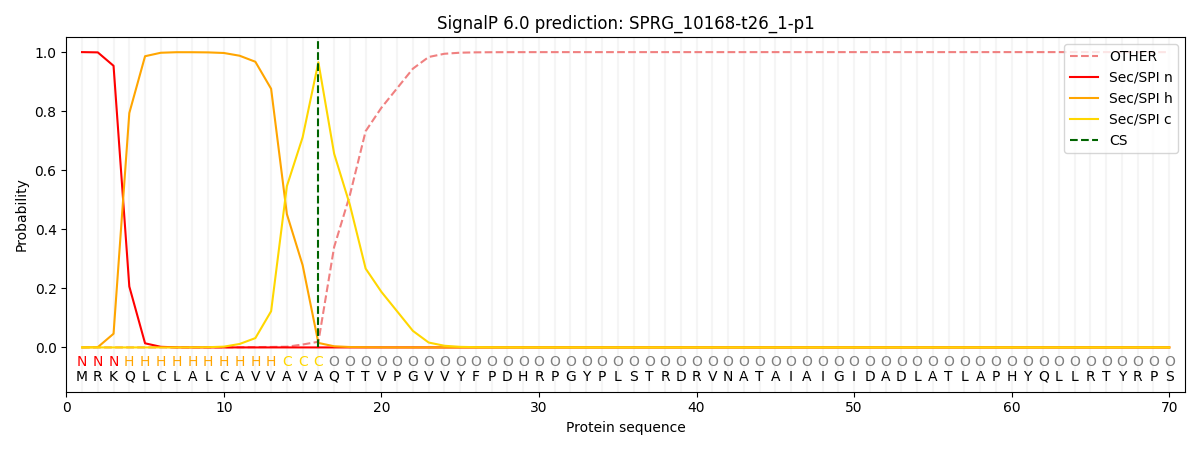
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000748 | 0.999214 | CS pos: 16-17. Pr: 0.9656 |