You are browsing environment: FUNGIDB
CAZyme Information: SPRG_09933-t26_1-p1
You are here: Home > Sequence: SPRG_09933-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_09933-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.109:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT105 | 77 | 203 | 3.9e-35 | 0.9236641221374046 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400627 | DUF1736 | 2.40e-06 | 235 | 293 | 5 | 63 | Domain of unknown function (DUF1736). This domain of unknown function is found in various hypothetical metazoan proteins. |
| 276809 | TPR | 1.09e-04 | 386 | 447 | 23 | 88 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 0.002 | 398 | 476 | 1 | 77 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 0.006 | 433 | 538 | 3 | 91 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.56e-68 | 6 | 523 | 16 | 575 | |
| 2.90e-67 | 6 | 523 | 16 | 575 | |
| 5.32e-65 | 15 | 523 | 129 | 687 | |
| 5.32e-65 | 15 | 523 | 129 | 687 | |
| 1.09e-64 | 5 | 523 | 11 | 570 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.46e-67 | 6 | 523 | 16 | 575 | Protein O-mannosyl-transferase TMTC3 OS=Mus musculus OX=10090 GN=Tmtc3 PE=1 SV=2 |
|
| 1.95e-65 | 5 | 523 | 11 | 570 | Protein O-mannosyl-transferase TMTC3 OS=Homo sapiens OX=9606 GN=TMTC3 PE=1 SV=2 |
|
| 8.30e-58 | 3 | 418 | 22 | 502 | Protein O-mannosyl-transferase TMTC4 OS=Homo sapiens OX=9606 GN=TMTC4 PE=1 SV=2 |
|
| 1.08e-56 | 3 | 418 | 21 | 502 | Protein O-mannosyl-transferase TMTC4 OS=Mus musculus OX=10090 GN=Tmtc4 PE=2 SV=1 |
|
| 1.75e-50 | 9 | 437 | 28 | 558 | Protein O-mannosyl-transferase TMTC1 OS=Drosophila melanogaster OX=7227 GN=CG31690 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.467135 | 0.532874 | CS pos: 16-17. Pr: 0.3160 |
TMHMM Annotations download full data without filtering help
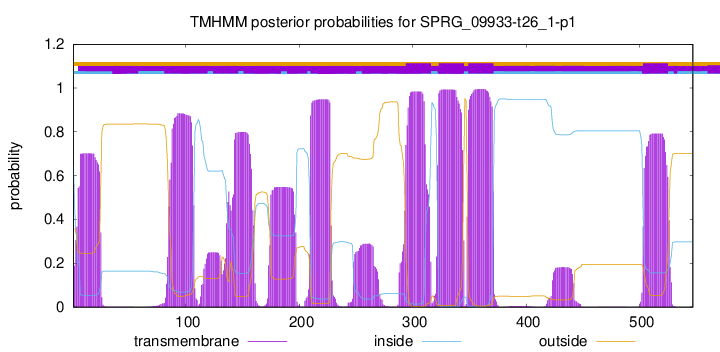
| Start | End |
|---|---|
| 294 | 316 |
| 323 | 345 |
| 349 | 371 |
| 503 | 525 |
