You are browsing environment: FUNGIDB
SPRG_07813-t26_1-p1
Basic Information
help
Species
Saprolegnia parasitica
Lineage
Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica
CAZyme ID
SPRG_07813-t26_1-p1
CAZy Family
GH2
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
244
KK583219|CGC7 26790.96
10.0706
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_SparasiticaCBS223-65
20435
695850
314
20121
Gene Location
Family
Start
End
Evalue
family coverage
PL7
38
223
1.7e-18
0.8457943925233645
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
400923
Alginate_lyase2
2.55e-15
24
223
1
222
Alginate lyase. Alginate lyases are enzymes that degrade the linear polysaccharide alignate. They cleave the glycosidic linkage of alignate through a beta-elimination reaction. This family forms an all beta fold and is different to all alpha fold of pfam05426.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
3.18e-89
6
239
3
274
8.46e-14
12
220
17
238
5.67e-13
6
220
9
236
6.37e-13
16
237
638
876
2.03e-12
17
224
73
297
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.51e-12
15
224
53
269
Alginate lyase AlgAT5 from Polysaccharide Lyase family 7 [Defluviitalea]
more
SPRG_07813-t26_1-p1 has no Swissprot hit.
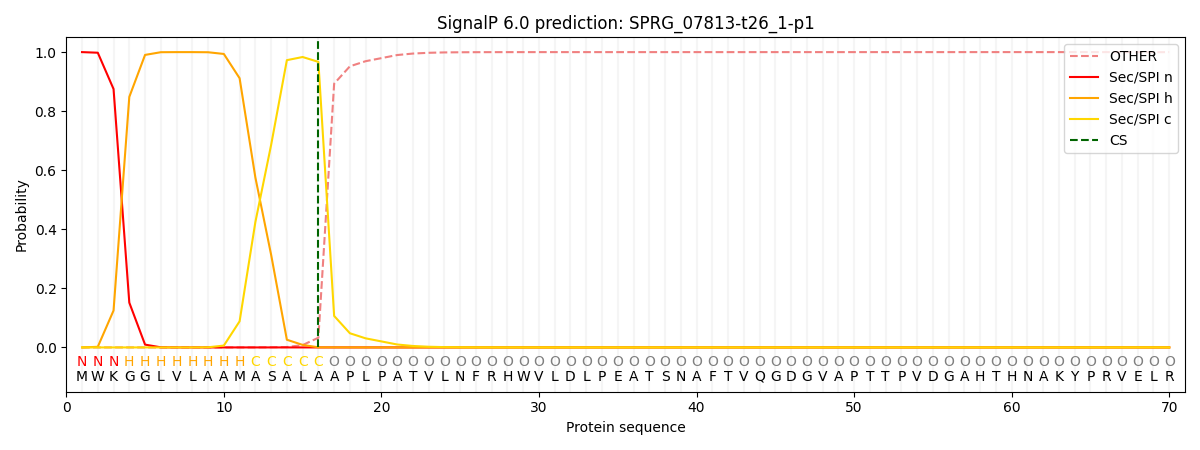
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000525
0.999445
CS pos: 16-17. Pr: 0.9668
There is no transmembrane helices in SPRG_07813-t26_1-p1.