You are browsing environment: FUNGIDB
CAZyme Information: SPRG_04092-t26_1-p1
You are here: Home > Sequence: SPRG_04092-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
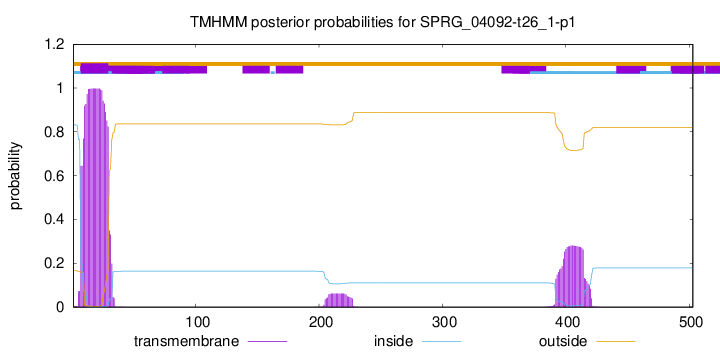
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_04092-t26_1-p1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT61 | 302 | 414 | 4.5e-19 | 0.8907563025210085 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398330 | DUF563 | 1.55e-22 | 296 | 441 | 77 | 206 | Protein of unknown function (DUF563). Family of uncharacterized proteins. |
| 226845 | COG4421 | 1.18e-08 | 342 | 437 | 232 | 324 | Capsular polysaccharide biosynthesis protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.48e-58 | 187 | 464 | 25 | 309 | |
| 4.48e-58 | 187 | 464 | 25 | 309 | |
| 9.35e-11 | 232 | 440 | 20 | 222 | |
| 3.48e-10 | 145 | 437 | 128 | 431 | |
| 4.93e-10 | 275 | 437 | 294 | 447 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.61e-06 | 241 | 437 | 267 | 451 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Drosophila melanogaster OX=7227 GN=Eogt PE=1 SV=1 |
|
| 3.78e-06 | 192 | 440 | 246 | 508 | Beta-1,2-xylosyltransferase RCN11 OS=Oryza sativa subsp. japonica OX=39947 GN=RCN11 PE=1 SV=1 |
|
| 8.73e-06 | 186 | 441 | 233 | 477 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Gallus gallus OX=9031 GN=EOGT PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
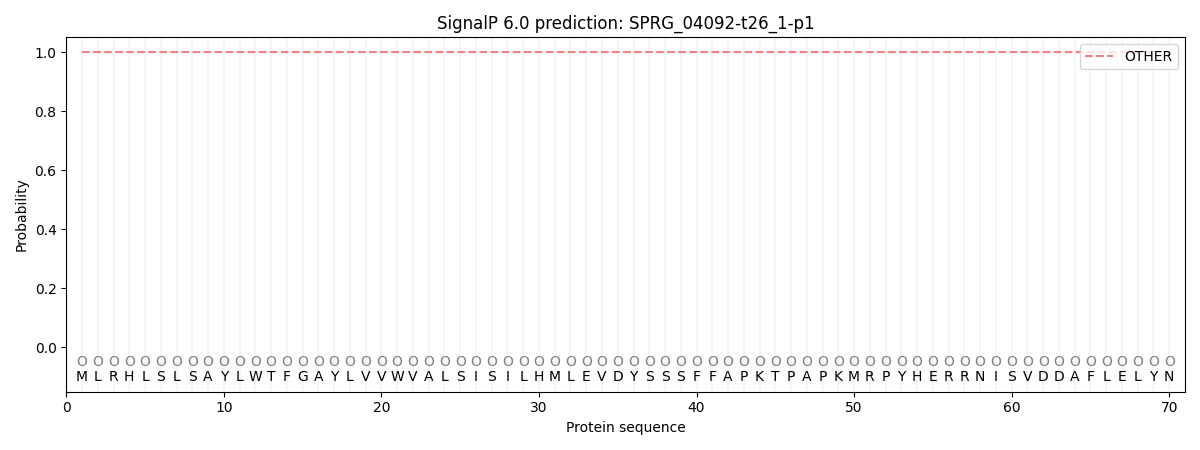
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000004 |