You are browsing environment: FUNGIDB
CAZyme Information: SPRG_00039-t26_1-p1
You are here: Home > Sequence: SPRG_00039-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Saprolegnia parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica | |||||||||||
| CAZyme ID | SPRG_00039-t26_1-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 239068 | Peptidase_C1A | 3.17e-65 | 127 | 334 | 4 | 210 | Peptidase C1A subfamily (MEROPS database nomenclature); composed of cysteine peptidases (CPs) similar to papain, including the mammalian CPs (cathepsins B, C, F, H, L, K, O, S, V, X and W). Papain is an endopeptidase with specific substrate preferences, primarily for bulky hydrophobic or aromatic residues at the S2 subsite, a hydrophobic pocket in papain that accommodates the P2 sidechain of the substrate (the second residue away from the scissile bond). Most members of the papain subfamily are endopeptidases. Some exceptions to this rule can be explained by specific details of the catalytic domains like the occluding loop in cathepsin B which confers an additional carboxydipeptidyl activity and the mini-chain of cathepsin H resulting in an N-terminal exopeptidase activity. Papain-like CPs have different functions in various organisms. Plant CPs are used to mobilize storage proteins in seeds. Parasitic CPs act extracellularly to help invade tissues and cells, to hatch or to evade the host immune system. Mammalian CPs are primarily lysosomal enzymes with the exception of cathepsin W, which is retained in the endoplasmic reticulum. They are responsible for protein degradation in the lysosome. Papain-like CPs are synthesized as inactive proenzymes with N-terminal propeptide regions, which are removed upon activation. In addition to its inhibitory role, the propeptide is required for proper folding of the newly synthesized enzyme and its stabilization in denaturing pH conditions. Residues within the propeptide region also play a role in the transport of the proenzyme to lysosomes or acidified vesicles. Also included in this subfamily are proteins classified as non-peptidase homologs, which lack peptidase activity or have missing active site residues. |
| 395062 | Peptidase_C1 | 2.91e-62 | 126 | 335 | 4 | 212 | Papain family cysteine protease. |
| 214761 | Pept_C1 | 9.01e-51 | 126 | 334 | 4 | 175 | Papain family cysteine protease. |
| 240310 | PTZ00200 | 1.88e-45 | 128 | 338 | 239 | 447 | cysteine proteinase; Provisional |
| 185513 | PTZ00203 | 5.51e-41 | 78 | 319 | 76 | 326 | cathepsin L protease; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.37e-211 | 11 | 573 | 11 | 585 | |
| 2.24e-185 | 11 | 564 | 6 | 558 | |
| 9.52e-183 | 7 | 582 | 9 | 580 | |
| 3.44e-51 | 343 | 570 | 305 | 544 | |
| 1.35e-44 | 341 | 571 | 250 | 493 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.90e-42 | 125 | 337 | 5 | 218 | The 2.1 Angstrom Structure Of A Cysteine Protease With Proline Specificity From Ginger Rhizome, Zingiber Officinale [Zingiber officinale],1CQD_B The 2.1 Angstrom Structure Of A Cysteine Protease With Proline Specificity From Ginger Rhizome, Zingiber Officinale [Zingiber officinale],1CQD_C The 2.1 Angstrom Structure Of A Cysteine Protease With Proline Specificity From Ginger Rhizome, Zingiber Officinale [Zingiber officinale],1CQD_D The 2.1 Angstrom Structure Of A Cysteine Protease With Proline Specificity From Ginger Rhizome, Zingiber Officinale [Zingiber officinale] |
|
| 1.16e-41 | 126 | 335 | 4 | 215 | Ficin A [Ficus carica],4YYQ_B Ficin A [Ficus carica] |
|
| 6.03e-41 | 126 | 338 | 5 | 221 | The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm [Ricinus communis],1S4V_B The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm [Ricinus communis] |
|
| 1.72e-40 | 127 | 335 | 5 | 216 | Actinidin from Actinidia arguta planch (Sarusashi) [Actinidia arguta] |
|
| 2.06e-40 | 126 | 340 | 7 | 230 | Crystal structure of recombinant barley cysteine endoprotease B isoform 2 (EP-B2) in complex with leupeptin [Hordeum vulgare],2FO5_B Crystal structure of recombinant barley cysteine endoprotease B isoform 2 (EP-B2) in complex with leupeptin [Hordeum vulgare],2FO5_C Crystal structure of recombinant barley cysteine endoprotease B isoform 2 (EP-B2) in complex with leupeptin [Hordeum vulgare],2FO5_D Crystal structure of recombinant barley cysteine endoprotease B isoform 2 (EP-B2) in complex with leupeptin [Hordeum vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.40e-43 | 6 | 334 | 12 | 337 | Senescence-specific cysteine protease SAG39 OS=Oryza sativa subsp. japonica OX=39947 GN=SAG39 PE=2 SV=2 |
|
| 5.49e-43 | 89 | 340 | 119 | 379 | Cysteine protease 1 OS=Oryza sativa subsp. japonica OX=39947 GN=CP1 PE=2 SV=2 |
|
| 6.09e-43 | 6 | 334 | 12 | 337 | Senescence-specific cysteine protease SAG39 OS=Oryza sativa subsp. indica OX=39946 GN=OsI_14861 PE=3 SV=1 |
|
| 1.23e-42 | 89 | 338 | 99 | 355 | Cysteine protease XCP1 OS=Arabidopsis thaliana OX=3702 GN=XCP1 PE=1 SV=1 |
|
| 1.77e-42 | 89 | 334 | 105 | 356 | Oryzain beta chain OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0670200 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
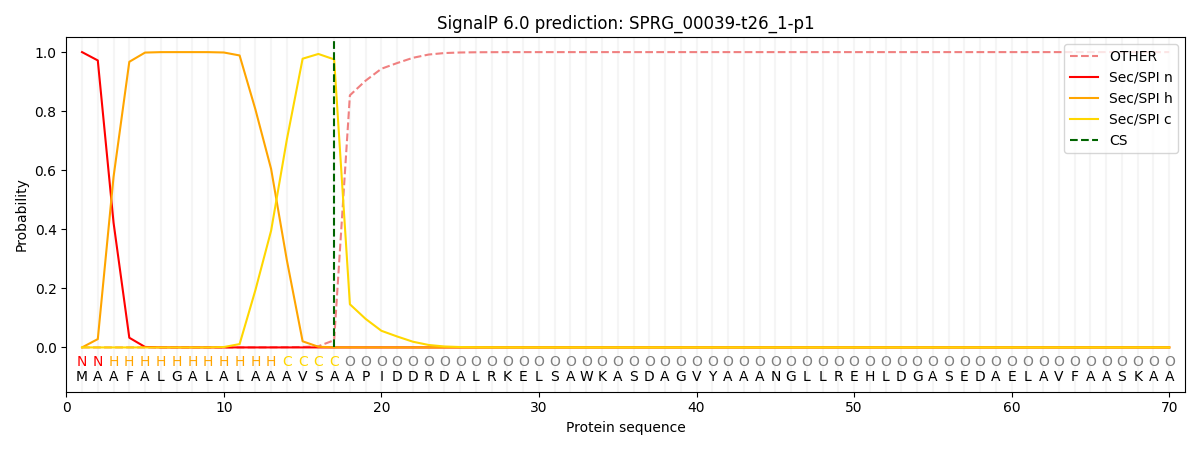
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000346 | 0.999633 | CS pos: 17-18. Pr: 0.9754 |
