You are browsing environment: FUNGIDB
CAZyme Information: SPBR_08521-t41_1-p1
You are here: Home > Sequence: SPBR_08521-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
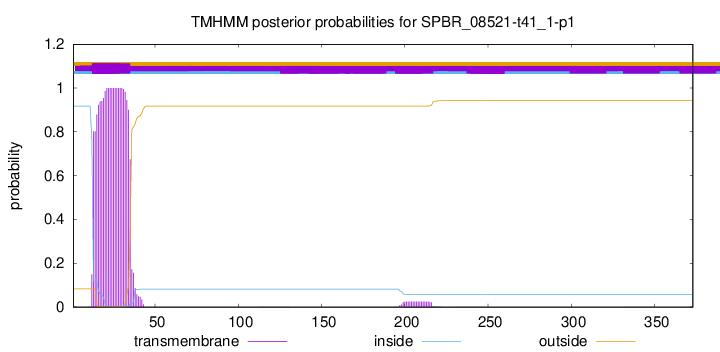
TMHMM annotations
Basic Information help
| Species | Sporothrix brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix brasiliensis | |||||||||||
| CAZyme ID | SPBR_08521-t41_1-p1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | mannan polymerase complexes MNN9 subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:2 | 2.4.1.257:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 75 | 339 | 6.4e-102 | 0.9813432835820896 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 7.28e-156 | 74 | 339 | 1 | 265 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 224137 | GT2 | 0.001 | 199 | 372 | 74 | 221 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.24e-204 | 1 | 373 | 1 | 372 | |
| 2.08e-203 | 1 | 368 | 1 | 367 | |
| 1.49e-198 | 1 | 373 | 1 | 374 | |
| 1.32e-197 | 1 | 371 | 1 | 370 | |
| 1.32e-197 | 1 | 371 | 1 | 370 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.00e-113 | 75 | 373 | 6 | 302 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.53e-112 | 75 | 373 | 96 | 392 | Mannan polymerase complexes subunit MNN9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN9 PE=1 SV=3 |
|
| 7.73e-110 | 75 | 373 | 73 | 368 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
|
| 3.11e-104 | 78 | 373 | 50 | 337 | Mannan polymerase complex subunit mnn9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnn9 PE=3 SV=1 |
|
| 7.59e-62 | 94 | 373 | 98 | 374 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 8.70e-51 | 14 | 373 | 11 | 357 | Mannan polymerase II complex ANP1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ANP1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
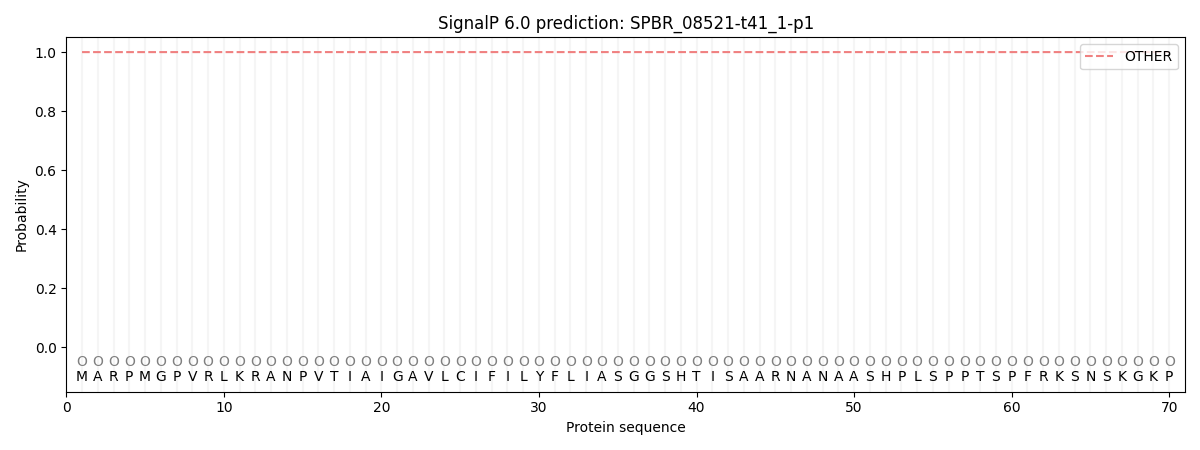
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000021 | 0.000003 |