You are browsing environment: FUNGIDB
CAZyme Information: SPBR_05081-t41_1-p1
You are here: Home > Sequence: SPBR_05081-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporothrix brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix brasiliensis | |||||||||||
| CAZyme ID | SPBR_05081-t41_1-p1 | |||||||||||
| CAZy Family | GH45 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 23 | 328 | 3.8e-96 | 0.9893617021276596 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 4.54e-56 | 39 | 323 | 8 | 272 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 7.74e-13 | 46 | 327 | 62 | 364 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.53e-212 | 17 | 388 | 19 | 389 | |
| 1.10e-209 | 17 | 388 | 19 | 390 | |
| 2.94e-208 | 17 | 388 | 18 | 388 | |
| 2.91e-205 | 6 | 388 | 4 | 384 | |
| 5.40e-192 | 1 | 389 | 1 | 387 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.91e-56 | 28 | 327 | 9 | 277 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
|
| 6.48e-55 | 28 | 327 | 11 | 279 | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger],5I79_B Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger] |
|
| 6.66e-55 | 28 | 327 | 12 | 280 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger],5I78_B Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
|
| 3.56e-50 | 18 | 326 | 1 | 279 | Crystal structure of an endoglucanase from Penicillium verruculosum [Talaromyces verruculosus] |
|
| 4.52e-50 | 18 | 326 | 10 | 288 | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose [Talaromyces verruculosus],5L9C_B Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose [Talaromyces verruculosus],5L9C_C Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose [Talaromyces verruculosus],5L9C_D Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose [Talaromyces verruculosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.18e-206 | 6 | 388 | 4 | 384 | Endoglucanase 1 OS=Robillarda sp. (strain Y-20) OX=72589 GN=eg 1 PE=1 SV=2 |
|
| 9.25e-55 | 28 | 327 | 39 | 307 | Probable endo-beta-1,4-glucanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=eglB PE=3 SV=1 |
|
| 3.89e-52 | 18 | 327 | 24 | 303 | Probable endo-beta-1,4-glucanase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglB PE=3 SV=1 |
|
| 1.07e-50 | 1 | 327 | 1 | 297 | Endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglA PE=1 SV=1 |
|
| 9.82e-50 | 25 | 327 | 35 | 307 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
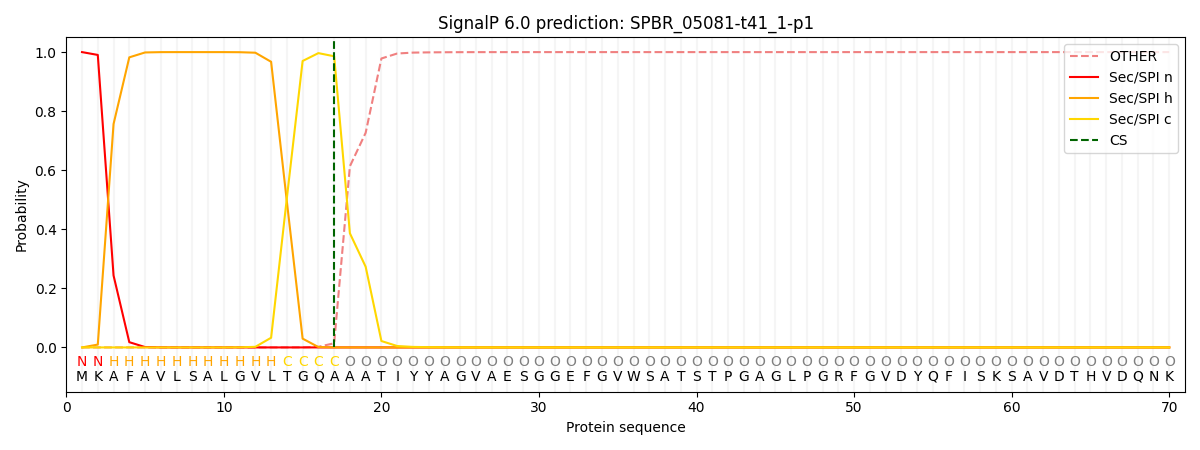
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000326 | 0.999659 | CS pos: 17-18. Pr: 0.9859 |
