You are browsing environment: FUNGIDB
CAZyme Information: SPBR_04834-t41_1-p1
You are here: Home > Sequence: SPBR_04834-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporothrix brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix brasiliensis | |||||||||||
| CAZyme ID | SPBR_04834-t41_1-p1 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | iron transport multicopper oxidase fet3 precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.10.3.2:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 101 | 295 | 3.6e-38 | 0.5519287833827893 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 259966 | CuRO_3_Fet3p | 5.05e-61 | 255 | 410 | 5 | 160 | The third Cupredoxin domain of multicopper oxidase Fet3p. Fet3p catalyzes the ferroxidase reaction, which couples the oxidation of Fe(II) to Fe(III) with the four-electron reduction of molecular oxygen to water. Fet3p is a type I membrane protein with the amino-terminal oxidase domain in the extracellular space and the carboxyl terminus in the cytoplasm. The periplasmic produced Fe(III) is transferred to the permease Ftr1p for import into the cytosol. The four copper ions are inserted post-translationally and are essential for catalytic activity, thus linking copper and iron homeostasis. Like other related multicopper oxidases (MCOs), Fet3p is composed of three cupredoxin domains that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 274555 | ascorbase | 1.85e-37 | 23 | 416 | 1 | 531 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 225043 | SufI | 7.13e-35 | 38 | 407 | 48 | 447 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 400194 | Cu-oxidase_2 | 5.51e-33 | 279 | 411 | 1 | 137 | Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognized by the pfam00394 model. |
| 259977 | CuRO_3_MCO_like_4 | 7.69e-33 | 264 | 406 | 10 | 164 | The third cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidize their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This subfamily of MCOs is composed of three cupredoxin domains. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.44e-182 | 21 | 460 | 22 | 541 | |
| 1.65e-178 | 7 | 460 | 4 | 541 | |
| 1.93e-168 | 10 | 460 | 9 | 549 | |
| 5.49e-168 | 16 | 460 | 15 | 549 | |
| 3.02e-162 | 11 | 460 | 10 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.03e-53 | 23 | 451 | 2 | 525 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 5.06e-31 | 27 | 449 | 7 | 501 | Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2],3KW7_B Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2] |
|
| 1.14e-25 | 5 | 450 | 3 | 518 | Crystal structure of laccases from Pycnoporus sanguineus, izoform I [Trametes sanguinea] |
|
| 3.83e-25 | 36 | 408 | 37 | 491 | Crystal structure of laccase from Lentinus sp. at 1.8 A resolution [Lentinus],3X1B_B Crystal structure of laccase from Lentinus sp. at 1.8 A resolution [Lentinus] |
|
| 4.38e-25 | 13 | 408 | 13 | 488 | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The fifth structure of the series with total exposition time 123 min. [Steccherinum murashkinskyi],5MHX_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The sixth structure of the series with total exposition time 153 min. [Steccherinum murashkinskyi],5MHY_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The seventh structure of the series with total exposition time 183 min. [Steccherinum murashkinskyi],5MHZ_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 8-th structure of the series with total exposition time 213 min. [Steccherinum murashkinskyi],5MI1_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 9-th structure of the series with total exposition time 243 min. [Steccherinum murashkinskyi],5MI2_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 10-th structure of the series with total exposition time 273 min. [Steccherinum murashkinskyi],5MIA_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 11-th structure of the series with total exposition time 303 min. [Steccherinum murashkinskyi],5MIB_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 12-th structure of the series with total exposition time 333 min. [Steccherinum murashkinskyi],5MIC_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 13-th structure of the series with total exposition time 363 min. [Steccherinum murashkinskyi],5MID_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 14-th structure of the series with total exposition time 393 min. [Steccherinum murashkinskyi],5MIE_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 15-th structure of the series with total exposition time 423 min. [Steccherinum murashkinskyi],5MIG_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 16-th structure of the series with total exposition time 453 min. The crystal was quick refreezing before this data collection. [Steccherinum murashkinskyi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-154 | 21 | 460 | 20 | 541 | Multicopper oxidase PfmaD OS=Pestalotiopsis fici (strain W106-1 / CGMCC3.15140) OX=1229662 GN=PfmaD PE=2 SV=1 |
|
| 6.30e-100 | 19 | 454 | 18 | 542 | Iron transport multicopper oxidase fetC OS=Epichloe festucae (strain E2368) OX=696363 GN=fetC PE=2 SV=1 |
|
| 1.17e-95 | 14 | 454 | 15 | 539 | Iron transport multicopper oxidase fetC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=fetC PE=2 SV=1 |
|
| 6.99e-91 | 5 | 455 | 4 | 548 | Iron transport multicopper oxidase FET3 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FET3 PE=2 SV=1 |
|
| 1.00e-72 | 21 | 443 | 18 | 535 | Multicopper oxidase abr1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=abr1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
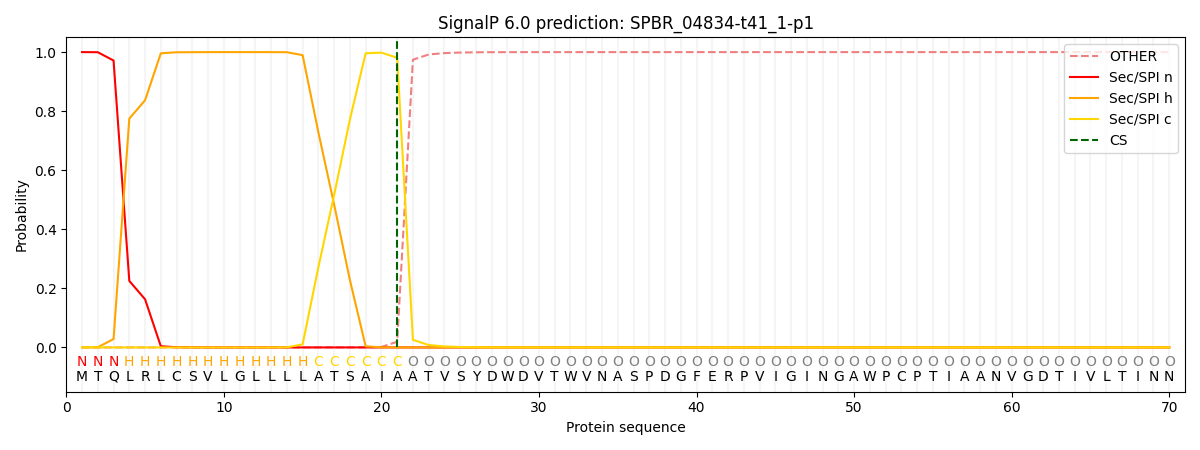
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000236 | 0.999736 | CS pos: 21-22. Pr: 0.9808 |
