You are browsing environment: FUNGIDB
CAZyme Information: SPBR_03397-t41_1-p1
You are here: Home > Sequence: SPBR_03397-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporothrix brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix brasiliensis | |||||||||||
| CAZyme ID | SPBR_03397-t41_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | c6 zinc finger domain containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.54:4 | 1.14.99.56:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA9 | 10 | 234 | 1.1e-57 | 0.9863636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397484 | Glyco_hydro_61 | 3.88e-86 | 33 | 239 | 9 | 211 | Glycosyl hydrolase family 61. Although weak endoglucanase activity has been demonstrated in several members of this family, they lack the clustered conserved catalytic acidic amino acids present in most glycoside hydrolases. Many members of this family lack measurable cellulase activity on their own, but enhance the activity of other cellulolytic enzymes. They are therefore unlikely to be true glycoside hydrolases. The subsrate-binding surface of this family is a flat Ig-like fold. |
| 410622 | LPMO_AA9 | 3.80e-85 | 22 | 247 | 1 | 216 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 9 (AA9). AA9 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs) involved in the cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C6. Activities include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54) and lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56). The family used to be called GH61 because weak endoglucanase activity had been demonstrated in some family members. |
| 213391 | fungal_TF_MHR | 1.01e-39 | 669 | 1048 | 1 | 363 | fungal transcription factor regulatory middle homology region. This domain is present in the large family of fungal zinc cluster transcription factors that contain an N-terminal GAL4-like C6 zinc binuclear cluster DNA-binding domain. Examples of members of this large fungal group are the following Saccharomyces cerevisiae transcription factors, GAL4, STB5, DAL81, CAT8, RDR1, HAL9, PUT3, PPR1, ASG1, RSF2, PIP2, as well as the C-terminal domain of the Cep3, a subunit of the yeast centromere-binding factor 3. It has been suggested that this region plays a regulatory role. |
| 397964 | Fungal_trans | 1.61e-15 | 678 | 901 | 1 | 219 | Fungal specific transcription factor domain. |
| 214903 | Fungal_trans | 4.01e-15 | 775 | 873 | 1 | 93 | Fungal specific transcription factor domain. This domain is found in a number of fungal transcription factors including transcriptional activator xlnR, yeast regulatory protein GAL4, and other transcription proteins regulating a variety of cellular and metabolic processes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.21e-94 | 11 | 268 | 7 | 265 | |
| 2.21e-94 | 11 | 268 | 7 | 265 | |
| 4.10e-93 | 11 | 268 | 7 | 265 | |
| 2.07e-92 | 19 | 268 | 15 | 265 | |
| 2.43e-75 | 16 | 256 | 16 | 257 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.71e-72 | 33 | 266 | 10 | 244 | Extended catalytic domain of Hypocrea jecorina LPMO 9A. [Trichoderma reesei QM6a],5O2X_A Extended catalytic domain of H. jecorina LPMO9A a.k.a EG4 [Trichoderma reesei QM6a] |
|
| 5.28e-72 | 33 | 250 | 10 | 229 | Chain A, Lytic polysaccharide monooxygenase [Talaromyces verruculosus] |
|
| 3.97e-70 | 33 | 248 | 10 | 227 | Thermoascus GH61 isozyme A [Thermoascus aurantiacus],2YET_B Thermoascus GH61 isozyme A [Thermoascus aurantiacus],3ZUD_A Thermoascus Gh61 Isozyme A [Thermoascus aurantiacus],7PU1_AAA Chain AAA, Gh61 isozyme a [Thermoascus aurantiacus],7PU1_BBB Chain BBB, Gh61 isozyme a [Thermoascus aurantiacus] |
|
| 1.24e-67 | 34 | 248 | 11 | 221 | Chain A, LPMO9F [Malbranchea cinnamomea] |
|
| 9.26e-65 | 33 | 248 | 10 | 227 | Chain AAA, Endoglucanase, putative [Aspergillus fischeri NRRL 181],7OVA_BBB Chain BBB, Endoglucanase, putative [Aspergillus fischeri NRRL 181],7OVA_CCC Chain CCC, Endoglucanase, putative [Aspergillus fischeri NRRL 181],7OVA_DDD Chain DDD, Endoglucanase, putative [Aspergillus fischeri NRRL 181] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.09e-71 | 33 | 265 | 29 | 263 | Polysaccharide monooxygenase Cel61a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Cel61a PE=1 SV=1 |
|
| 1.11e-70 | 11 | 266 | 11 | 265 | Endoglucanase-4 OS=Hypocrea jecorina OX=51453 GN=cel61a PE=1 SV=1 |
|
| 7.18e-50 | 51 | 247 | 51 | 247 | Endoglucanase-7 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cel61b PE=1 SV=3 |
|
| 1.54e-26 | 628 | 985 | 237 | 566 | Transcription factor lepE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=lepB PE=3 SV=1 |
|
| 1.09e-25 | 14 | 249 | 22 | 256 | Cellulose-growth-specific protein OS=Agaricus bisporus OX=5341 GN=cel1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
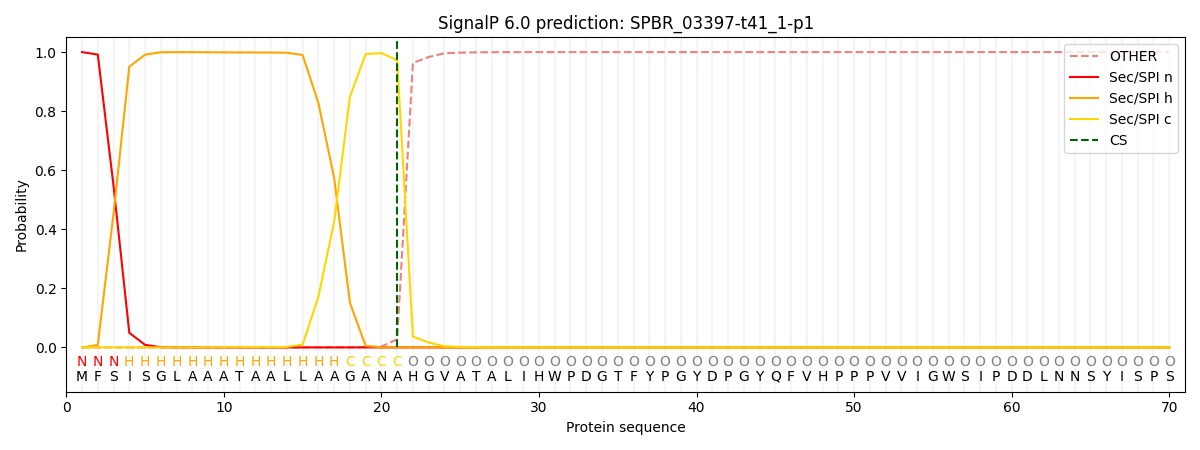
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000343 | 0.999634 | CS pos: 21-22. Pr: 0.9727 |
