You are browsing environment: FUNGIDB
CAZyme Information: SPBR_00400-t41_1-p1
You are here: Home > Sequence: SPBR_00400-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporothrix brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Ophiostomataceae; Sporothrix; Sporothrix brasiliensis | |||||||||||
| CAZyme ID | SPBR_00400-t41_1-p1 | |||||||||||
| CAZy Family | AA16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 29 | 331 | 7e-19 | 0.8716216216216216 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 5.86e-89 | 28 | 335 | 1 | 280 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 197609 | LysM | 1.98e-10 | 418 | 464 | 1 | 44 | Lysin motif. |
| 395573 | Glyco_hydro_18 | 2.19e-10 | 29 | 304 | 1 | 260 | Glycosyl hydrolases family 18. |
| 396179 | LysM | 4.51e-10 | 419 | 464 | 1 | 42 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| 212030 | LysM | 6.30e-10 | 417 | 464 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.64e-83 | 22 | 411 | 14 | 403 | |
| 1.48e-79 | 13 | 411 | 5 | 403 | |
| 1.63e-78 | 24 | 338 | 15 | 333 | |
| 1.63e-74 | 24 | 335 | 15 | 346 | |
| 2.79e-74 | 13 | 411 | 5 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.38e-59 | 30 | 337 | 3 | 307 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 6.57e-59 | 30 | 337 | 4 | 308 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 6.57e-59 | 30 | 337 | 4 | 308 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 4.70e-39 | 30 | 337 | 8 | 287 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 1.51e-35 | 28 | 325 | 1 | 268 | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR [Hevea brasiliensis],1LLO_A Chain A, HEVAMINE [Hevea brasiliensis],2HVM_A Hevamine A At 1.8 Angstrom Resolution [Hevea brasiliensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.67e-64 | 24 | 348 | 24 | 351 | Endochitinase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=ctcA PE=2 SV=1 |
|
| 1.05e-54 | 24 | 348 | 25 | 347 | Endochitinase A1 OS=Neosartorya fumigata OX=746128 GN=chiA1 PE=1 SV=1 |
|
| 5.99e-54 | 24 | 348 | 25 | 347 | Endochitinase A1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chiA1 PE=3 SV=1 |
|
| 1.92e-52 | 26 | 349 | 27 | 349 | Endochitinase 2 OS=Coccidioides posadasii (strain C735) OX=222929 GN=CTS2 PE=3 SV=2 |
|
| 2.50e-52 | 26 | 349 | 27 | 349 | Endochitinase 2 OS=Coccidioides immitis (strain RS) OX=246410 GN=CTS2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
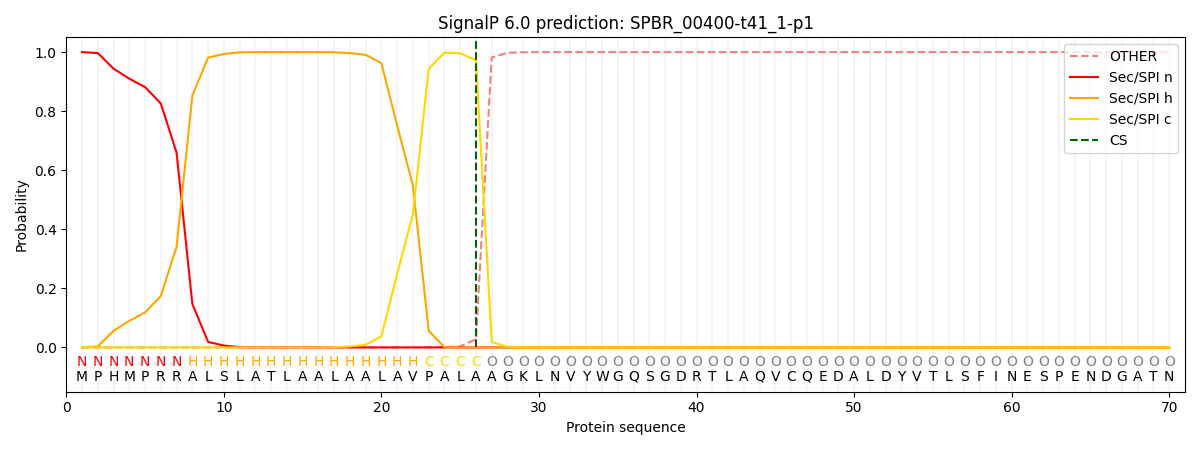
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000373 | 0.999601 | CS pos: 26-27. Pr: 0.9722 |
