You are browsing environment: FUNGIDB
CAZyme Information: SOCG_02535-t26_1-p1
You are here: Home > Sequence: SOCG_02535-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
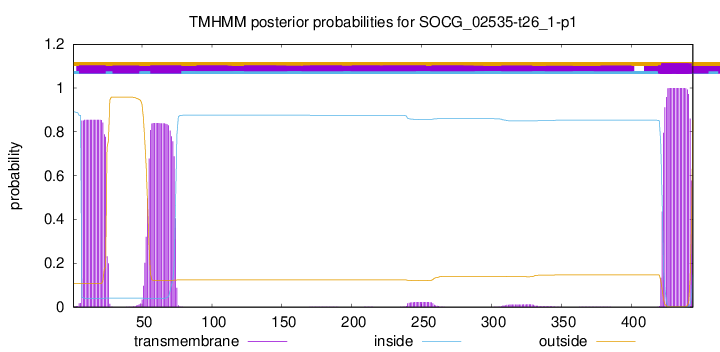
TMHMM annotations
Basic Information help
| Species | Schizosaccharomyces octosporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Schizosaccharomycetes; ; Schizosaccharomycetaceae; Schizosaccharomyces; Schizosaccharomyces octosporus | |||||||||||
| CAZyme ID | SOCG_02535-t26_1-p1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | mannan endo-1,6-alpha-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 31 | 384 | 2.3e-86 | 0.9301675977653632 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.21e-134 | 31 | 374 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 2.97e-07 | 125 | 313 | 86 | 275 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.02e-207 | 30 | 444 | 28 | 442 | |
| 3.02e-207 | 30 | 444 | 28 | 442 | |
| 2.92e-128 | 7 | 404 | 5 | 407 | |
| 2.92e-128 | 7 | 404 | 5 | 407 | |
| 4.25e-123 | 9 | 441 | 8 | 444 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-77 | 29 | 404 | 33 | 425 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.37e-208 | 30 | 444 | 28 | 442 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
|
| 5.18e-129 | 7 | 404 | 5 | 407 | Putative mannan endo-1,6-alpha-mannosidase C970.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC970.02 PE=3 SV=1 |
|
| 7.55e-124 | 9 | 441 | 8 | 444 | Putative mannan endo-1,6-alpha-mannosidase C1198.06c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.06c PE=3 SV=1 |
|
| 6.65e-109 | 17 | 429 | 22 | 443 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
|
| 7.58e-109 | 9 | 430 | 9 | 437 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000246 | 0.999741 | CS pos: 21-22. Pr: 0.9785 |