You are browsing environment: FUNGIDB
CAZyme Information: SMR63396.1
You are here: Home > Sequence: SMR63396.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | SMR63396.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 215 | 411 | 6.3e-21 | 0.8105726872246696 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273828 | esterase_phb | 4.66e-41 | 217 | 430 | 1 | 211 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| 340881 | MFS_Tpo1_MDR_like | 2.91e-34 | 31 | 170 | 175 | 315 | Yeast Polyamine transporter 1 (Tpo1) and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This family is composed of fungal multidrug resistance (MDR) transporters including several proteins from Saccharomyces cerevisiae such as polyamine transporters 1-4 (Tpo1-4), quinidine resistance proteins 1-3 (Qdr1-3), dityrosine transporter 1 (Dtr1), fluconazole resistance protein 1 (Flr1), and protein HOL1. MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. For example, Flr1 confers resistance to the azole derivative fluconazole while Tpo1 confers resistance and adaptation to quinidine and ketoconazole. The polyamine transporters are involved in the detoxification of excess polyamines in the cytoplasm. Tpo1-like MDR transporters belong to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 402228 | Esterase_phd | 7.25e-21 | 220 | 418 | 6 | 200 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| 226040 | LpqC | 4.17e-20 | 168 | 457 | 3 | 277 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 226584 | COG4099 | 3.13e-06 | 301 | 342 | 261 | 302 | Predicted peptidase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 470 | 1 | 470 | |
| 0.0 | 1 | 470 | 1 | 470 | |
| 0.0 | 1 | 465 | 1 | 512 | |
| 0.0 | 1 | 470 | 1 | 501 | |
| 1.12e-174 | 170 | 469 | 1 | 300 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.86e-96 | 201 | 470 | 3 | 275 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.38e-97 | 193 | 470 | 23 | 304 | Acetylxylan esterase A OS=Aspergillus awamori OX=105351 GN=axeA PE=1 SV=2 |
|
| 3.78e-97 | 195 | 470 | 24 | 303 | Probable acetylxylan esterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=axeA PE=3 SV=1 |
|
| 5.35e-97 | 195 | 470 | 24 | 303 | Acetylxylan esterase A OS=Aspergillus ficuum OX=5058 GN=axeA PE=1 SV=1 |
|
| 8.52e-97 | 197 | 468 | 29 | 303 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 3.45e-96 | 173 | 468 | 5 | 304 | Probable acetylxylan esterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000022 | 0.000022 |
TMHMM Annotations download full data without filtering help
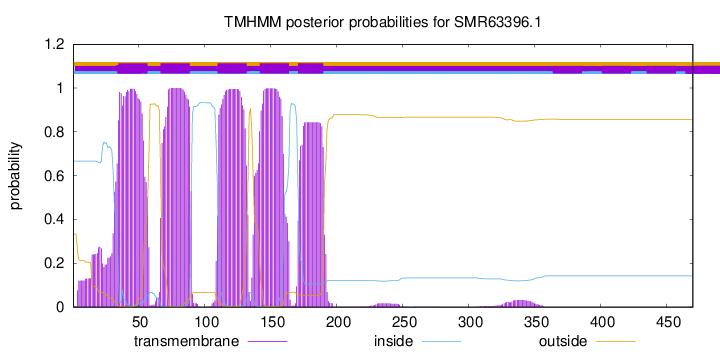
| Start | End |
|---|---|
| 35 | 57 |
| 67 | 89 |
| 110 | 132 |
| 142 | 164 |
| 171 | 190 |
