You are browsing environment: FUNGIDB
CAZyme Information: SMR62701.1
You are here: Home > Sequence: SMR62701.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | SMR62701.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA8 | 568 | 723 | 9.2e-16 | 0.8681318681318682 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 187688 | CDH_like_cytochrome | 2.25e-49 | 564 | 719 | 15 | 168 | Heme-binding cytochrome domain of fungal cellobiose dehydrogenases. Cellobiose dehydrogenase (CellobioseDH or CDH) is an extracellular fungal oxidoreductase that degrades both lignin and cellulose. Specifically, CDHs oxidize cellobiose, cellodextrins, and lactose to corresponding lactones, utilizing a variety of electron acceptors. Class-II CDHs are monomeric hemoflavoenzymes that are comprised of a b-type cytochrome domain linked to a large flavodehydrogenase domain. The cytochrome domain of CDH and related enzymes, which this model describes, folds as a beta sandwich and complexes a heme molecule. It is found at the N-terminus of this family of enzymes, and belongs to the DOMON domain superfamily, a ligand-interacting motif found in all three kingdoms of life. |
| 176490 | Cyt_b561_FRRS1_like | 2.91e-39 | 711 | 901 | 1 | 187 | Eukaryotic cytochrome b(561), including the FRRS1 gene product. Cytochrome b(561), as found in eukaryotes, similar to and including the human FRRS1 gene product (ferric-chelate reductase 1), also called SDR-2 (stromal cell-derived receptor 2). This family comprises a variety of domain architectures, many of which contain dopamine beta-monooxygenase (DOMON) domains. The protein might act as a ferric-chelate reductase, catalyzing the reduction of Fe(3+) to Fe(2+), such as associated with the transport of iron from the endosome to the cytoplasm. It is assumed that this protein uses ascorbate as the electron donor. Belongs to the cytochrome b(561) family, which are secretory vesicle-specific electron transport proteins. Cytochromes b(561) are integral membrane proteins that bind two heme groups non-covalently, and may have six alpha-helical trans-membrane segments. |
| 395859 | Cu2_monooxygen | 3.19e-29 | 216 | 337 | 1 | 129 | Copper type II ascorbate-dependent monooxygenase, N-terminal domain. The N and C-terminal domains of members of this family adopt the same PNGase F-like fold. |
| 406418 | CDH-cyt | 3.80e-27 | 566 | 723 | 16 | 174 | Cytochrome domain of cellobiose dehydrogenase. CDH-cyt is the cytochrome domain, at the N-terminus, of cellobiose dehydrogenase. CDH-cyt folds as a beta sandwich with the topology of the antibody Fab V(H) domain and binds iron. The haem iron is ligated by Met83 and His181 in UniProtKB:Q01738. |
| 397669 | Cu2_monoox_C | 1.35e-26 | 360 | 498 | 4 | 144 | Copper type II ascorbate-dependent monooxygenase, C-terminal domain. The N and C-terminal domains of members of this family adopt the same PNGase F-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.95e-104 | 560 | 918 | 22 | 392 | |
| 4.95e-104 | 560 | 918 | 22 | 392 | |
| 4.01e-96 | 561 | 925 | 22 | 395 | |
| 3.80e-89 | 560 | 916 | 23 | 389 | |
| 1.38e-88 | 570 | 918 | 25 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.50e-26 | 188 | 498 | 150 | 464 | Human dopamine beta-hydroxylase [Homo sapiens],4ZEL_B Human dopamine beta-hydroxylase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.95e-37 | 54 | 501 | 38 | 475 | DBH-like monooxygenase protein 2 homolog OS=Danio rerio OX=7955 GN=moxd2 PE=2 SV=2 |
|
| 1.01e-36 | 48 | 501 | 25 | 483 | DBH-like monooxygenase protein 1 OS=Homo sapiens OX=9606 GN=MOXD1 PE=1 SV=1 |
|
| 5.78e-36 | 48 | 501 | 25 | 483 | DBH-like monooxygenase protein 1 OS=Mus musculus OX=10090 GN=Moxd1 PE=1 SV=1 |
|
| 3.32e-35 | 47 | 501 | 26 | 485 | DBH-like monooxygenase protein 1 homolog OS=Danio rerio OX=7955 GN=moxd1 PE=2 SV=2 |
|
| 1.39e-34 | 59 | 498 | 32 | 477 | Tyramine beta-hydroxylase OS=Caenorhabditis briggsae OX=6238 GN=tbh-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
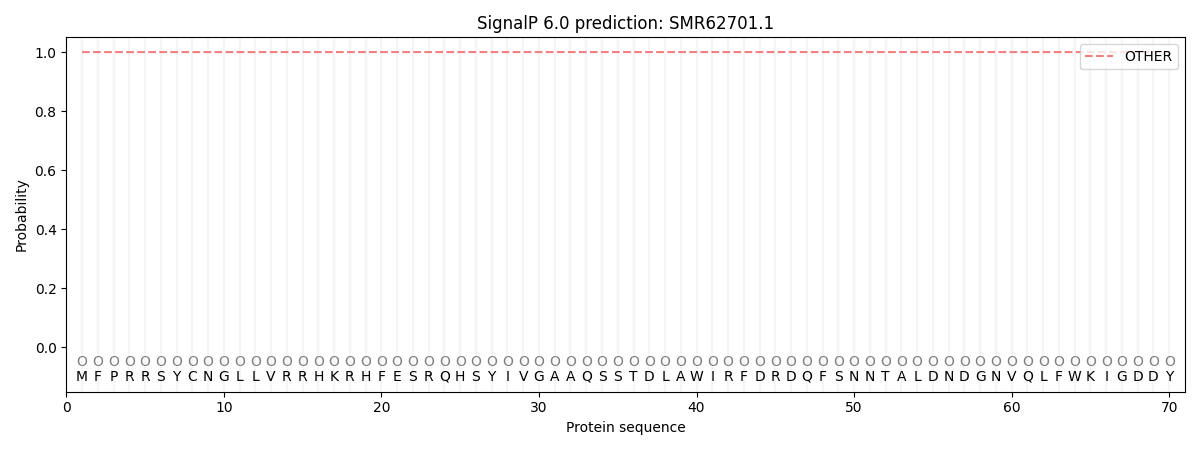
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000042 | 0.000000 |
TMHMM Annotations download full data without filtering help
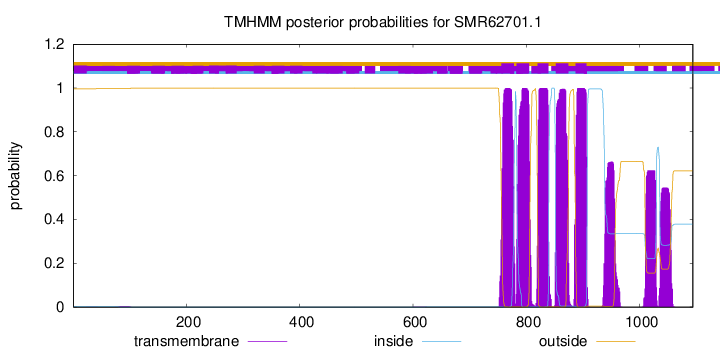
| Start | End |
|---|---|
| 756 | 778 |
| 783 | 805 |
| 820 | 839 |
| 852 | 874 |
| 887 | 906 |
