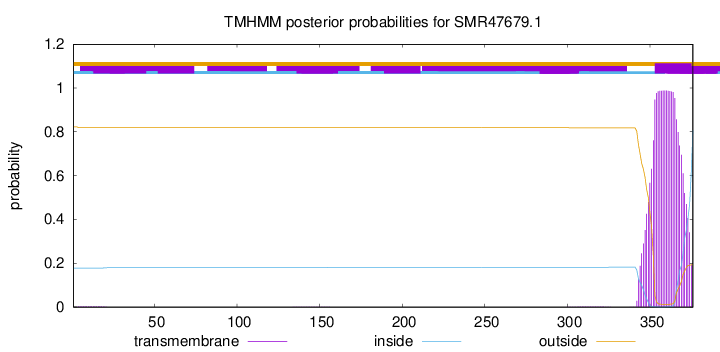You are browsing environment: FUNGIDB
CAZyme Information: SMR47679.1
You are here: Home > Sequence: SMR47679.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | SMR47679.1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH152 | 41 | 315 | 2.8e-61 | 0.9953703703703703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395248 | Thaumatin | 5.31e-87 | 41 | 315 | 1 | 211 | Thaumatin family. |
| 185757 | TLP-PA | 4.57e-58 | 36 | 314 | 1 | 219 | allergenic/antifungal thaumatin-like proteins: plant and animal homologs. This subfamily is represented by the thaumatin-like proteins (TLPs), Cherry Allergen Pru Av 2 TLP, Peach PpAZ44 TLP (a propylene-induced TLP in abscission), the Caenorhabditis elegans thaumatin family member (thn-6), and other plant and animal homologs. TLPs are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Due to their inducible expression by environmental stresses such as pathogen/pest attack, drought and cold, plant TLPs are classified as the pathogenesis-related (PR) protein family 5 (PR5). Several members of the plant TLP family have been reported as food allergens from fruits (i.e., cherry, Pru av 2; bell pepper, Cap a1; tomatoes, Lyc e NP24) and pollen allergens from conifers (i.e., mountain cedar, Jun a 3; Arizona cypress, Cup a3; Japanese cedar, Cry j3). TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. It has been proposed that the antifungal activity of plant PR5 proteins relies on the strong electronegative character of this cleft. Some TLPs hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. TLPs within this subfamily contain 16 conserved Cys residues. |
| 128501 | THN | 2.76e-50 | 37 | 315 | 1 | 218 | Thaumatin family. The thaumatin family gathers proteins related to plant pathogenesis. The thaumatin family includes very basic members with extracellular and vacuolar localization. Thaumatin itsel is a potent sweet-tasting protein. Several members of this family display significant in vitro activity of inhibiting hyphal growth or spore germination of various fungi probably by a membrane permeabilizing mechanism. |
| 185754 | Thaumatin-like | 5.15e-48 | 37 | 314 | 1 | 157 | the sweet-tasting protein, thaumatin, and thaumatin-like proteins involved in host defense. This family is represented by the sweet-tasting protein thaumatin from the African berry Thaumatococcus daniellii and thaumatin-like proteins (TLPs) involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Plant TLPs are classified as pathogenesis-related (PR) protein family 5 (PR5), their expression is induced by environmental stresses such as pathogen/pest attack, drought and cold. TLPs included in this family are such proteins as zeamatin, found in high concentrations in cereal seeds; osmotin, a salt-induced protein in osmotically stressed plants; and PpAZ44, a propylene-induced TLP in abscission of young fruit. Several members of the plant TLP family have been reported as food allergens from fruits (i.e., cherry, Pru av 2; bell pepper, Cap a1; tomatoes, Lyc e NP24) and pollen allergens from conifers (i.e., mountain cedar, Jun a 3; Arizona cypress, Cup a3; Japanese cedar, Cry j3). Thaumatin and TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. It has been proposed that the antifungal activity of plant PR5 proteins relies on the strong electronegative character of this cleft. Some TLPs hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. Most TLPs contain 16 conserved Cys residues. A deletion within the third domain (domain II) of the Triticum aestivum thaumatin-like xylanase inhibitor is observed, thus, only 10 conserved Cys residues are present within this smaller TLP and similar homologs. |
| 185756 | TLP-P | 1.06e-30 | 37 | 150 | 1 | 107 | thaumatin and allergenic/antifungal thaumatin-like proteins: plant homologs. This subfamily is represented by the sweet-tasting protein thaumatin from the African berry Thaumatococcus daniellii, allergenic/antifungal Thaumatin-like proteins (TLPs), and related plant proteins. TLPs are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Plant TLPs are classified as pathogenesis-related (PR) protein family 5 (PR5), their expression is induced by environmental stresses such as pathogen/pest attack, drought and cold. TLPs in this subfamily include such proteins as zeamatin, found in high concentrations in cereal seeds, and osmotin, a salt-induced protein in osmotically stressed plants. Several members of the plant TLP family have been reported as food allergens from fruits (i.e., cherry, Pru av 2; bell pepper, Cap a1; tomatoes, Lyc e NP24) and pollen allergens from conifers (i.e., mountain cedar, Jun a 3; Arizona cypress, Cup a3; Japanese cedar, Cry j3). Thaumatin and TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. It has been proposed that the antifungal activity of plant PR5 proteins relies on the strong electronegative character of this cleft. IgE-binding epitopes of mountain Cedar (Juniperus ashei) allergen Jun a 3, which interact with pooled IgE from patients suffering allergenic response to this allergen, were mainly located on the helical domain II; the best-conserved IgE-binding epitope predicted for TLPs corresponds to this region. Some TLPs hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. Most TLPs contain 16 conserved Cys residues. A deletion within the third domain (domain II) of the Triticum aestivum thaumatin-like xylanase inhibitor is observed, thus, only 10 conserved Cys residues are present within this smaller TLP and similar homologs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.51e-137 | 5 | 325 | 4 | 324 | |
| 3.66e-124 | 22 | 350 | 30 | 344 | |
| 6.37e-119 | 37 | 343 | 19 | 338 | |
| 1.26e-115 | 16 | 323 | 37 | 351 | |
| 2.18e-104 | 30 | 323 | 4 | 303 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.92e-37 | 36 | 315 | 2 | 222 | High resolution structure of Mal d 2, the thaumatin like food allergen from apple [Malus domestica] |
|
| 4.03e-34 | 37 | 315 | 3 | 222 | High resolution structure of a cherry allergen Pru av 2 [Prunus avium] |
|
| 2.40e-33 | 39 | 315 | 5 | 201 | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins [Vitis vinifera] |
|
| 8.89e-33 | 39 | 315 | 5 | 200 | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7A [Musa acuminata] |
|
| 2.15e-30 | 39 | 317 | 6 | 206 | Crystal Structure of Osmotin, an antifungal laticifer protein [Calotropis procera] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.85e-40 | 32 | 315 | 30 | 256 | Thaumatin-like protein 1 OS=Arabidopsis thaliana OX=3702 GN=TLP1 PE=2 SV=1 |
|
| 2.20e-39 | 39 | 337 | 29 | 272 | PR5-like receptor kinase OS=Arabidopsis thaliana OX=3702 GN=PR5K PE=1 SV=1 |
|
| 2.77e-39 | 36 | 315 | 24 | 244 | Thaumatin-like protein 1 OS=Pyrus pyrifolia OX=3767 GN=TL1 PE=1 SV=1 |
|
| 4.88e-39 | 41 | 315 | 29 | 238 | Pathogenesis-related thaumatin-like protein 3.5 OS=Cryptomeria japonica OX=3369 PE=1 SV=1 |
|
| 3.59e-38 | 39 | 315 | 28 | 239 | Pathogenesis-related protein 5 OS=Arabidopsis thaliana OX=3702 GN=At1g75040 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
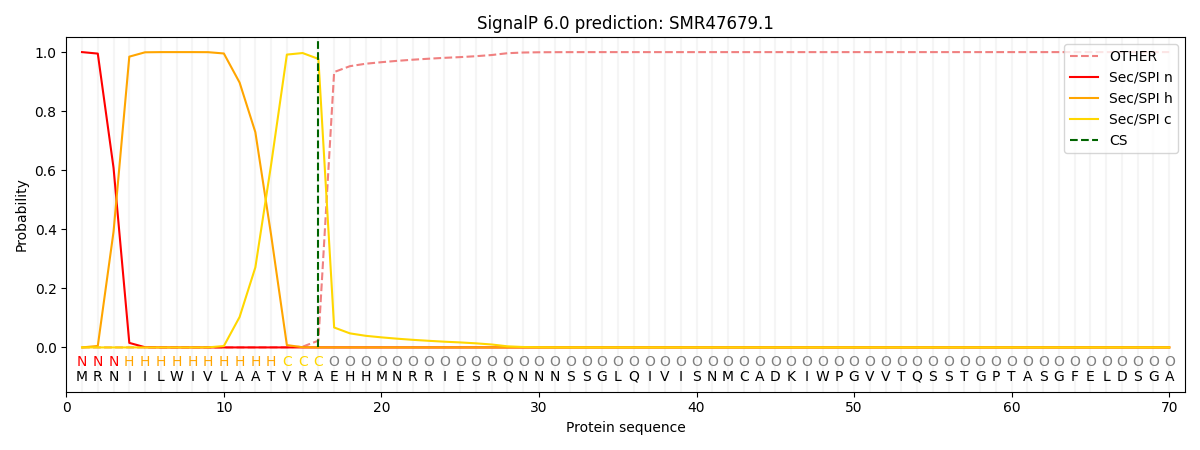
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000267 | 0.999703 | CS pos: 16-17. Pr: 0.9769 |