You are browsing environment: FUNGIDB
CAZyme Information: SMAC_06957-t26_1-p1
You are here: Home > Sequence: SMAC_06957-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
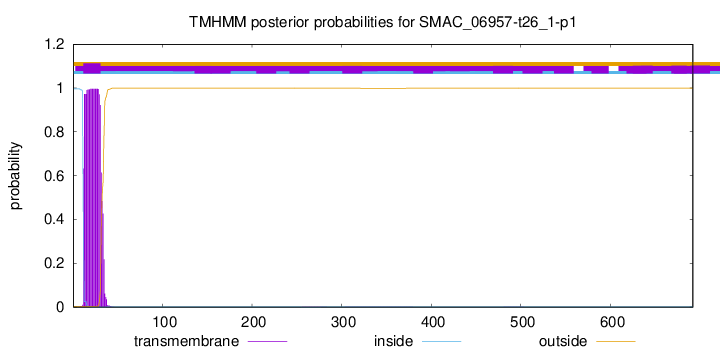
TMHMM annotations
Basic Information help
| Species | Sordaria macrospora | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Sordaria; Sordaria macrospora | |||||||||||
| CAZyme ID | SMAC_06957-t26_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 34 | 235 | 8.2e-66 | 0.9842931937172775 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237057 | PRK12323 | 0.002 | 290 | 506 | 376 | 598 | DNA polymerase III subunit gamma/tau. |
| 223021 | PHA03247 | 0.005 | 290 | 465 | 2722 | 2910 | large tegument protein UL36; Provisional |
| 411344 | gliding_GltG | 0.008 | 387 | 464 | 102 | 178 | adventurous gliding motility protein GltG. GltG proteins, including the founding member MXAN_4867 from Myxococcus xanthus, occur in certain delta-proteobacteria and are involved in adventurous gliding (A-)motility. GltG has an N-terminal forkhead-associated (FHA) domain domain, often associated with signal transduction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.68e-121 | 25 | 550 | 31 | 485 | |
| 3.07e-58 | 24 | 255 | 8 | 236 | |
| 5.93e-58 | 24 | 255 | 8 | 236 | |
| 1.11e-57 | 24 | 255 | 8 | 235 | |
| 8.34e-57 | 24 | 244 | 8 | 226 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.71e-49 | 34 | 229 | 1 | 199 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000830 | 0.999134 | CS pos: 33-34. Pr: 0.9647 |