You are browsing environment: FUNGIDB
CAZyme Information: SHO80146.1
You are here: Home > Sequence: SHO80146.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
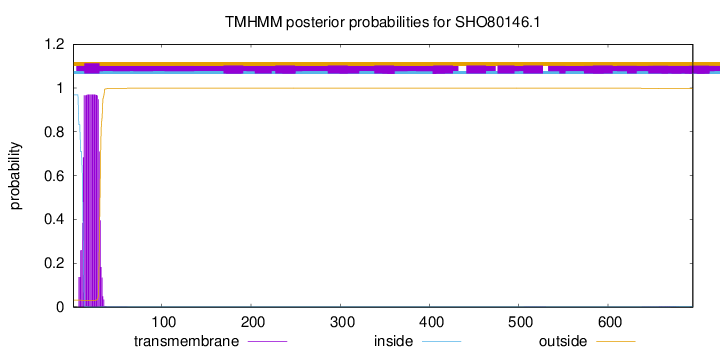
TMHMM annotations
Basic Information help
| Species | Malassezia sympodialis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Malasseziomycetes; ; Malasseziaceae; Malassezia; Malassezia sympodialis | |||||||||||
| CAZyme ID | SHO80146.1 | |||||||||||
| CAZy Family | GT59 | |||||||||||
| CAZyme Description | Similar to S.cerevisiae protein FET5 (Multicopper oxidase) [Source:UniProtKB/TrEMBL;Acc:A0A1M8ACE6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 100 | 674 | 1.6e-94 | 0.9804469273743017 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 1.74e-70 | 84 | 679 | 2 | 523 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 259953 | CuRO_2_MCO_like_1 | 3.20e-69 | 250 | 418 | 1 | 163 | The second cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This family of MCOs is composed of three cupredoxin domains that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 2 of 3-domain MCOs has lost the ability to bind copper. |
| 177843 | PLN02191 | 6.73e-65 | 84 | 679 | 24 | 546 | L-ascorbate oxidase |
| 259977 | CuRO_3_MCO_like_4 | 4.60e-59 | 506 | 679 | 2 | 166 | The third cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidize their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This subfamily of MCOs is composed of three cupredoxin domains. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 215324 | PLN02604 | 4.52e-56 | 82 | 694 | 23 | 561 | oxidoreductase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 14 | 694 | 13 | 695 | |
| 0.0 | 14 | 694 | 13 | 695 | |
| 6.38e-302 | 7 | 694 | 6 | 680 | |
| 1.28e-301 | 7 | 694 | 6 | 680 | |
| 3.14e-136 | 74 | 691 | 52 | 634 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.28e-53 | 84 | 694 | 4 | 538 | Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1AOZ_B Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1ASO_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASO_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASP_A X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASP_B X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASQ_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASQ_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo] |
|
| 1.46e-46 | 50 | 679 | 33 | 542 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
|
| 2.69e-46 | 89 | 691 | 10 | 482 | Chain A, LACCASE 1 [Coprinopsis cinerea] |
|
| 2.74e-46 | 89 | 691 | 10 | 482 | Chain A, Laccase [Coprinopsis cinerea] |
|
| 3.43e-46 | 85 | 691 | 5 | 482 | Chain A, Laccase [Trametes versicolor],1KYA_B Chain B, Laccase [Trametes versicolor],1KYA_C Chain C, Laccase [Trametes versicolor],1KYA_D Chain D, Laccase [Trametes versicolor] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.83e-60 | 65 | 685 | 41 | 566 | Laccase-2 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC2 PE=3 SV=2 |
|
| 6.64e-58 | 50 | 684 | 25 | 565 | Laccase-1 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=LAC1 PE=1 SV=1 |
|
| 3.22e-57 | 80 | 694 | 35 | 574 | L-ascorbate oxidase OS=Cucumis sativus OX=3659 PE=1 SV=1 |
|
| 2.09e-55 | 50 | 681 | 25 | 562 | Laccase-1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC1 PE=1 SV=1 |
|
| 3.97e-54 | 88 | 679 | 28 | 543 | L-ascorbate oxidase OS=Arabidopsis thaliana OX=3702 GN=AAO PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
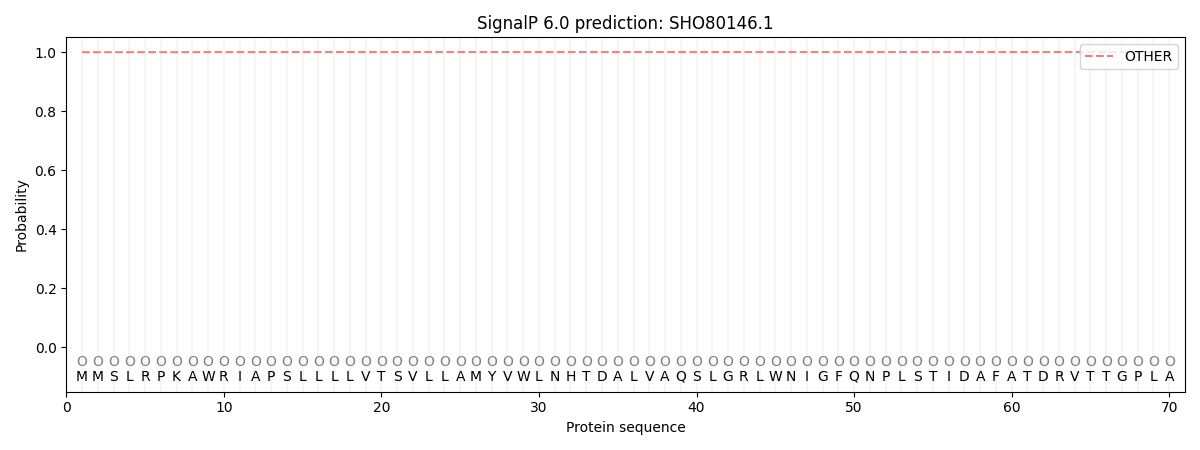
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999559 | 0.000467 |