You are browsing environment: FUNGIDB
CAZyme Information: SHO77403.1
You are here: Home > Sequence: SHO77403.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Malassezia sympodialis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Malasseziomycetes; ; Malasseziaceae; Malassezia; Malassezia sympodialis | |||||||||||
| CAZyme ID | SHO77403.1 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | Similar to S.cerevisiae protein KRE5 (Protein required for beta-1,6 glucan biosynthesis) [Source:UniProtKB/TrEMBL;Acc:A0A1M8A4L3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:29 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT24 | 1245 | 1492 | 4.2e-121 | 0.9959677419354839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408203 | Glyco_transf_24 | 0.0 | 1245 | 1512 | 1 | 268 | Glucosyltransferase 24. This is the catalytic domain found in UDP-glucose:glycoprotein glucosyltransferase (UGGT). This domain belongs to glucosyltransferase 24 family (GT24) A-type domain. The GT domain displays the expected glycosyltransferase type A (GT-A) fold. |
| 133054 | GT8_HUGT1_C_like | 7.79e-159 | 1245 | 1492 | 1 | 248 | The C-terminal domain of HUGT1-like is highly homologous to the GT 8 family. C-terminal domain of glycoprotein glucosyltransferase (UGT). UGT is a large glycoprotein whose C-terminus contains the catalytic activity. This catalytic C-terminal domain is highly homologous to Glycosyltransferase Family 8 (GT 8) and contains the DXD motif that coordinates donor sugar binding, characteristic for Family 8 glycosyltransferases. GT 8 proteins are retaining enzymes based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. The non-catalytic N-terminal portion of the human UTG1 (HUGT1) has been shown to monitor the protein folding status and activate its glucosyltransferase activity. |
| 132996 | Glyco_transf_8 | 4.84e-54 | 1245 | 1486 | 1 | 242 | Members of glycosyltransferase family 8 (GT-8) are involved in lipopolysaccharide biosynthesis and glycogen synthesis. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. GT-8 comprises enzymes with a number of known activities: lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, glycogenin glucosyltransferase, and N-acetylglucosaminyltransferase. GT-8 enzymes contains a conserved DXD motif which is essential in the coordination of a catalytic divalent cation, most commonly Mn2+. |
| 408201 | Thioredoxin_14 | 2.67e-31 | 452 | 709 | 1 | 245 | Thioredoxin-like domain. This is the third out of four TRXL(thioredoxin-like) domains found in UDP-glucose:glycoprotein glucosyltransferase (UGGT). |
| 399437 | UDP-g_GGTase | 7.22e-27 | 1101 | 1208 | 1 | 107 | UDP-glucose:Glycoprotein Glucosyltransferase. UDP-g_GGTase is an important, central component of the QC system in the ER for checking that glycoproteins are folded correctly. This QC prevents incorrectly folded glycoproteins from leaving the ER. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1530 | 1 | 1530 | |
| 0.0 | 4 | 1530 | 5 | 1527 | |
| 0.0 | 49 | 1530 | 1 | 1479 | |
| 5.37e-283 | 3 | 1516 | 13 | 1626 | |
| 2.12e-276 | 25 | 1516 | 34 | 1627 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.10e-202 | 107 | 1518 | 78 | 1453 | Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],5MZO_A Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],5N2J_A Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],5N2J_B Chain B, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],6TRF_A Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 3.45e-201 | 107 | 1518 | 78 | 1453 | Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 9.35e-201 | 107 | 1518 | 78 | 1453 | Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 7.83e-168 | 107 | 1454 | 71 | 1382 | Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],6TS8_B Chain B, UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 3.64e-161 | 818 | 1518 | 491 | 1219 | Chain A, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],6TS2_B Chain B, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],6TS2_C Chain C, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495],6TS2_D Chain D, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein [Thermochaetoides thermophila DSM 1495] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.20e-184 | 200 | 1530 | 175 | 1448 | UDP-glucose:glycoprotein glucosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gpt1 PE=1 SV=2 |
|
| 2.90e-154 | 107 | 1530 | 96 | 1516 | UDP-glucose:glycoprotein glucosyltransferase 2 OS=Homo sapiens OX=9606 GN=UGGT2 PE=1 SV=4 |
|
| 5.06e-154 | 116 | 1521 | 106 | 1516 | UDP-glucose:glycoprotein glucosyltransferase OS=Drosophila melanogaster OX=7227 GN=Uggt PE=1 SV=2 |
|
| 3.21e-150 | 180 | 1516 | 182 | 1572 | UDP-glucose:glycoprotein glucosyltransferase OS=Arabidopsis thaliana OX=3702 GN=UGGT PE=1 SV=1 |
|
| 4.00e-148 | 145 | 1529 | 147 | 1539 | UDP-glucose:glycoprotein glucosyltransferase 1 OS=Homo sapiens OX=9606 GN=UGGT1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
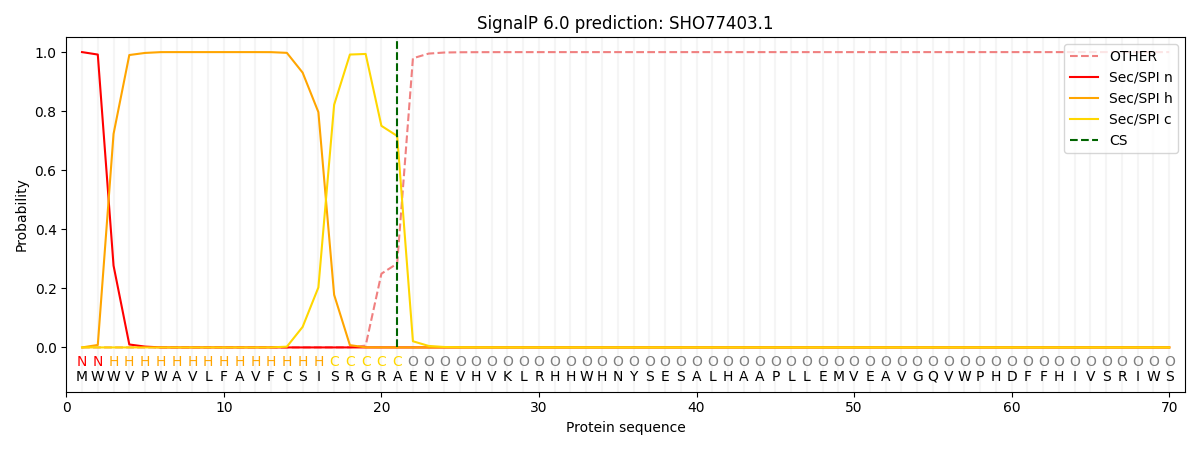
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000207 | 0.999769 | CS pos: 21-22. Pr: 0.7161 |
